Abstract
Malignant astrocytoma includes anaplastic astrocytoma (grade III) and glioblastoma (grade IV). Among them, glioblastoma is the most common primary brain tumor with dismal responses to all therapeutic modalities. We performed a large-scale, genome-wide microRNA (miRNA) (n=756) expression profiling of 26 glioblastoma, 13 anaplastic astrocytoma and 7 normal brain samples with an aim to find deregulated miRNA in malignant astrocytoma. We identified several differentially regulated miRNAs between these groups, which could differentiate glioma grades and normal brain as recognized by PCA. More importantly, we identified a most discriminatory 23-miRNA expression signature, by using PAM, which precisely distinguished glioblastoma from anaplastic astrocytoma with an accuracy of 95%. The differential expression pattern of nine miRNAs was further validated by real-time RT-PCR on an independent set of malignant astrocytomas (n=72) and normal samples (n=7). Inhibition of two glioblastoma-upregulated miRNAs (miR-21 and miR-23a) and exogenous overexpression of two glioblastoma-downregulated miRNAs (miR-218 and miR-219-5p) resulted in reduced soft agar colony formation but showed varying effects on cell proliferation and chemosensitivity. Thus we have identified the miRNA expression signature for malignant astrocytoma, in particular glioblastoma, and showed the miRNA involvement and their importance in astrocytoma development.
Similar content being viewed by others
Main
MicroRNAs (miRNAs) are endogenous non-coding small RNAs, which negatively regulate gene expression either by degrading specific mRNA or inhibiting translation. miRNAs have been linked to variety of cancers, in which miRNAs were shown to act like either oncogenes or tumor suppressors.1 Classification of multiple cancers on the basis of expression pattern of 217 miRNAs was found to be more accurate than using expression pattern of 16 000 mRNAs.2 Furthermore, 52.5% of miRNA genes are found in cancer-associated genomic regions or at fragile sites, minimal loss of heterozygosity regions, minimal regions of amplification or at common break point regions.3 Consequently, it is very important to identify the differentially regulated miRNAs in cancer and identify the target genes to advance our understanding of the mechanism how miRNAs modulate gene function forming regulatory networks in cancer.
Astrocytoma, the tumor of astrocytic glial cells, is the most common type of central nervous system (CNS) neoplasms, accounting for more than 60% of all primary brain tumors.4 Diffuse infiltrating astrocytomas are divided into low-grade diffuse astrocytoma (DA; grade II), anaplastic astrocytoma (AA; grade III) and glioblastoma (GBM; grade IV) in the order of increasing malignancy.5 Patients with diffuse astrocytoma have the best prognosis with a median survival time of 6 to 8 years after surgical intervention as compared with anaplastic astrocytoma and glioblastoma, which are classified as ‘High-grade or malignant astrocytomas’. The treatment protocols and length of survival are distinctly different between anaplastic astrocytoma and glioblastoma patients.6, 7 The median survival time for anaplastic astrocytoma patients is 2 to 3 years whereas patients with glioblastoma have a median survival of less than 1 year.8 As histology-based classification is highly subjective, there is a need for more robust histology-independent molecular classifiers.9
Glioblastomas have been further subdivided into primary glioblastoma (pGBM) and secondary glioblastoma (sGBM) subtypes on the basis of clinical and molecular profile. Primary glioblastoma presents in an acute de novo manner with no evidence of an antecedent lower grade tumor. By contrast, secondary glioblastoma results from the progressive malignant transformation of a diffuse astrocytoma or anaplastic astrocytoma.10 Remarkably, despite their distinct clinical profiles, pGBMs and sGBMs are reflected by an equally poor prognosis when adjusted for patient age.10 However, although these glioblastoma subtypes achieve a common clinical endpoint, recent studies have identified markedly different transcriptional patterns and DNA copy number variations between them.11, 12
In view of the above and to obtain a histology-independent miRNA expression signature to differentiate glioblastoma from anaplastic astrocytoma tumors, we have analyzed the expression of 756 miRNAs by microarray in 13 anaplastic astrocytoma and 29 glioblastoma tumor samples. In addition to identifying grade-specific miRNA signature, we found a 23-miRNA expression signature that precisely differentiated glioblastoma from anaplastic astrocytoma with an accuracy rate of 95%. Besides performing real-time RT-PCR validation of selected miRNAs, we also analyzed the functional importance of four of these miRNAs.
Materials and methods
Cell Lines and Tumor Samples
The glioma cell lines SVG, U138, U251, U343, U373, U87, LN18 and LN229 were grown in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% FBS. Tumor samples were collected from patients who were operated at National Institute of Mental Health and Neurosciences (NIMHANS) and Sri Sathya Sai Institute of Higher Medical Sciences (SSSIHMS), Bangalore, India. A portion of the non-dominant anterior temporal cortex resected during surgery for intractable epilepsy served as normal control brain sample. Tissues were freshly received from the neurosurgical operating rooms, bisected and one half was placed in RNA later (Ambion Inc., USA), stored at −70°C and used for RNA isolation. The other half was fixed in 10% buffered neutral formalin, processed for paraffin sections and was used for histopathology and immunohistochemistry (IHC). The study has been scrutinized and approved by the ethics committee of the two clinical centers, NIMHANS and SSIHMS, and patient consent was obtained before initiation of the study as per the Institute Ethical Committee guidelines and approval. A total of 125 samples of malignant astrocytoma and control brain tissue were used in this study. For microarray hybridization, a set of 46 samples comprising 13 anaplastic astrocytoma, 13 secondary glioblastoma, 13 primary glioblastoma and 7 normal brain controls were used. For RT-PCR validation of selected miRNAs, we used an independent set of 51 glioblastoma, 21 anaplastic astrocytoma and 7 normal brain controls.
RNA Isolation and miRNA Expression Profiling
Total RNA isolation from the frozen tissue or cell lines and quantitation were performed as described before.12 Total RNA (1 μg) from sample and reference was labeled with Hy3™ and Hy5™ fluorescent label, respectively, using the miRCURY™ LNA Array power labeling kit (Exiqon, Denmark). The common reference RNA was prepared by mixing equal quantity of RNA from all the samples including normal and tumor. The Hy3-labeled samples and an Hy5-labeled reference RNA sample were mixed pair-wise and hybridized to the miRCURY LNA array version 10.0 (Exiqon), which contains capture probes targeting all miRNAs for all species registered in the miRBASE version 10.0 at the Sanger Institute. The hybridization was performed using a Tecan HS4800 hybridization station (Tecan, Austria). The miRCURY LNA array microarray slides were scanned using the G2565BA Microarray Scanner System (Agilent, USA) and image analysis was performed using the ImaGene 7.0 software (BioDiscovery, USA). The quantified signals were background-corrected (Normexp with offset value 10)13 and normalized using the global Lowess (LOcally WEighted Scatterplot Smoothing) regression algorithm.
Data analysis
The median ratio of Hy3/Hy5 intensity for replicative spots for each miRNA was obtained after normalization. The values for Hy3/Hy5 ratio were log2-transformed. The difference in log2 ratio for each miRNA (subtracting the log2 ratio of a given sample from the average log2 ratio of normal samples) was considered in subsequent analyses. miRNAs having values in at least 70% of the samples in each group were considered for further analysis. Data were analyzed by Significance Analysis of Microarrays (SAM) to identify differentially regulated miRNAs between any two groups. In SAM, P-values were obtained from permutation tests (1000 permutations in each analysis). The significant genes that were identified by significance analysis of microarrays were subjected to Hierarchical Clustering Analysis (HCA) to identify the pattern of expression across different groups. TMeV software version 4.0 was used to perform SAM and HCA.
Class prediction analysis was performed by Prediction Analysis of Microarray (PAM) using the PAM package in the R software (version 2.7.1). The probability was calculated in the test data set by 10-fold cross-validation.14 Principal Component Analysis (PCA) was performed using R package (version 2.7.1).
TaqMan Quantitative Real-Time PCR of the miRNAs
We used TaqMan real-time PCR method (Applied Biosystems) to measure the level of mature miRNAs. miRNA-specific cDNA was made using a specific stem–loop primer and a diluted reverse transcription product from 10 ng total RNA was used for each real-time PCR reaction. Real-time PCR was performed using the ABI 7900HT real-time PCR system. The PCR conditions were as follows: 95°C for 10 min, and then 15 s at 95°C and 1 min at 60°C for 40 cycles. U6 RNA was used as endogenous control. Delta Ct values were used for the analysis. Statistical significance was tested by Mann–Whitney test using the GraphPad PRISM software.
Histopathology and IHC
Histological sections of normal brain and tumor tissues were examined by light microscopy using hematoxylin and eosin preparation. Tumor sections of diffusely infiltrating astrocytomas were graded using the WHO grading scheme. Paraffin sections (4 μm) from tumor tissue and control samples were collected on silane-coated slides for IHC. The antibodies used mainly for the purpose of sub-classifying GBM cases were p53 (monoclonal: DO-7; Biogenix, diluted to 1:200) and EGFR (monoclonal: E-30; Biogenix, diluted to 1:50). For p53, antigen retrieval was performed by heat treatment of the deparaffinized sections in a microwave oven for 25 to 35 min at 700 W in citrate buffer (10 mmol/l, pH 6.0). For EGFR staining, the sections were pretreated with Tris-EDTA (pH 9.0) at 600 W for 30 min. All sections were further treated with methanol and 5% hydrogen peroxide to block endogenous peroxidase followed by washes with PBS buffer (pH 7.6). Skimmed milk powder (5%) was used to block background staining for 45 min. The sections were incubated with the primary antibody overnight at 4°C. This was followed by incubation with the supersensitive non-biotin horseradish peroxidase detection system (QD440-XAK; Biogenex). 3,3-Diaminobenzidine (Sigma) was used as a chromogenic substrate. Brain tumor samples previously characterized for overexpression of p53 and EGFR were used as positive controls. p53 and EGFR immunoreactivity was seen as nuclear and membrane cytoplasmic labeling, respectively. Glioblastomas were classified as primary and secondary, taking into consideration mainly the clinical profile of patients. EGFR and p53 expression status was used as an adjunct to segregate these groups. The mean age of patients with primary glioblastoma was 52 years and the mean duration of symptoms was 1 month. Uniform staining for EGFR was evident in 12/13 cases and six showed additional p53 expression. Among secondary glioblastomas, the mean age of the patients was 41 years and the mean duration of symptoms was 10 months. p53 immunoreactivity was uniformly evident in 12/13 cases and none showed EGFR overexpression.
Transfection of Cells with Anti-miR Inhibitors or Pre-miR Mimicks
The U138 cell line was used for the downregulation of miR-21 and miR-23a, and overexpression of miR-218 and miR-219-5p. For downregulation of overexpressed miRNAs, the Anti-miR-inhibitors and Cy-3-labeled Anti-miR-negative control were obtained from Ambion. For overexpression, the precursors of the corresponding miRNAs and Cy-3-labeled Pre-miR control were obtained from Ambion. Transfection was performed by adding the inhibitors/precursors at a concentration of 30 nM using the siPORT NeoFX transfection agent (Ambion) following the manufacturer's instructions. After transfection, RNA was isolated at the indicated time points and the overexpression/downregulation of miRNA was confirmed by assaying for the level of mature miRNA by TaqMan quantitative real-time PCR.
Proliferation and Chemosensitivity Assays
U138 cells were transfected in 24-well plates with miRNA-specific Pre-miR, Anti-miR and control precursor/inhibitors as described before. At different time intervals, the cells were trypsinized, suspended in PBS and viable cells were counted. For chemosensitivity assay, U138 cells were transfected with miRNA-specific Pre-miR, Anti-miR and control precursor/inhibitors as described before. The chemotherapeutic drugs were added at the indicated concentrations 36 h after transfection. The proportion of viable cells was measured by MTT assay.15
Anchorage-Independent Colony Formation Assay in Soft Agar
U138 cells were transfected with miRNA-specific Pre-miR, Anti-miR and control precursor/inhibitors as described before. After 36 h of transfection, the cells were harvested and plated for soft agar assay. Base agar was prepared by using 800 μl of 0.6% agar in complete medium in a six-well plate. After the base agar was set, agar solution was mixed with 5 × 103 U138 cells (final concentration 0.34%) and overlaid on the solidified base agar. The plates were incubated at 37°C for up to 2 weeks. Duplicate wells were prepared for each transfection. Colonies were counted and photographed.
Results
Differential Expression of miRNAs between Malignant Astrocytomas and Normal Brains
We performed a genome-wide miRNA expression profiling of 756 human miRNAs on 39 glioma samples (26 glioblastomas consisting of 13 primary glioblastomas and 13 secondary glioblastomas; 13 anaplastic astrocytomas) and 7 normal brain samples (see Supplementary Table S1). The data obtained from image analysis was filtered, normalized and log2-transformed before using for further analysis. To identify the significantly differentially regulated miRNAs between normal and astrocytoma samples, we analyzed a total of 287 miRNAs through SAM using the two-class option.16 At a false discovery rate (FDR) of 0.0%, we found 55 miRNAs to be upregulated and 29 miRNAs downregulated in malignant astrocytomas in comparison with normal brain samples. For visual appreciation of miRNA regulation, the data obtained from SAM was subjected to a supervised, one-way, average-linkage HCA with Euclidean distance as similarity metric, using the TMev software (Supplementary Figure S1 and Supplementary Table S2). We further performed PCA using a subset of SAM-identified significant miRNAs. Malignant astrocytoma samples (blue, purple and orange triangles) and normal brain (green triangle) samples separated clearly into two major clusters (Figure 1a). Furthermore, within malignant astrocytoma, glioblastomas (blue and purple triangles) and anaplastic astrocytomas (orange triangles) formed two sub-clusters (upper vs lower section, respectively). It is interesting to note that few of the tumor samples appeared to group with the grade different from that identified by histopathology. We have investigated the reasons for the discordance and the details are provided later. Interestingly, glioblastoma samples appeared to form one single cluster even though this group included 13 each of secondary glioblastoma (blue triangles) and primary glioblastoma (purple triangles).
Identification of tumor grade-specific miRNA. (a) PCA was performed using the expression values of a subset of SAM-identified differentially regulated miRNAs (n=47) between normal brain tissue samples (n=7) and malignant astrocytoma tumor samples (n=39) as shown in Supplementary Table S2. A scatter plot is generated using the first three principal components for each sample. The color of the samples is as indicated. (b–d) One-way hierarchical clustering of SAM-identified, grade-specific miRNAs that differentiate glioblastoma from anaplastic astrocytoma (b), progressive astrocytoma from de novo glioblastoma (c) and primary glioblastoma from secondary glioblastoma. (d) A dual-color code was used, with red and green indicating up- and downregulation, respectively.
Differentially Regulated miRNAs between Malignant Astrocytoma Grades
Next we focused on miRNA signature, which can differentiate one grade of astrocytoma from the other. On comparison of anaplastic astrocytoma and glioblastoma, SAM identified (FDR of 0.0%) a total of 67 miRNAs to be differentially regulated between these two groups (Figure 1b and Supplementary Table S3). Among these 67 differentially regulated miRNAs, there were four sub-clusters having an interesting pattern of expression. Cluster-A and B essentially consisted of miRNAs, which were upregulated and downregulated in both anaplastic astrocytoma and glioblastoma, with upregulation and downregulation reaching substantially high levels in glioblastoma in comparison with anaplastic astrocytoma. Cluster-C and D were more interesting because the expression levels of these miRNAs in anaplastic astrocytoma were either less than normal (cluster-C) or similar to normal (cluster-D) whereas they are highly upregulated in glioblastoma samples. To find unique differences between secondary glioblastoma and primary glioblastoma in miRNA expression signature, we performed the following comparisons. On comparison of secondary glioblastoma and primary glioblastoma independently with anaplastic astrocytoma (n=39; 13 secondary glioblastoma, 13 primary glioblastoma and 13 anaplastic astrocytoma), there were 68 and 76 miRNAs differentially regulated in secondary glioblastoma and primary glioblastoma, respectively, as compared with anaplastic astrocytoma at an FDR of 0.00% (Supplementary Figure S2A and B, and Supplementary Tables S4 and S5). Interestingly, among these differentially regulated miRNAs, 50 miRNAs were common to secondary glioblastoma and primary glioblastoma whereas there were 18 and 26 miRNAs unique to secondary glioblastoma and primary glioblastoma, respectively, as compared with anaplastic astrocytoma. When progressive astrocytoma samples (n=26; 13 anaplastic astrocytoma and 13 secondary glioblastoma) were compared against primary glioblastoma (de novo glioblastoma; n=13 primary glioblastoma), we found 21 miRNAs to be differentially regulated (Figure 1c and Supplementary Table S6). On comparison of secondary glioblastoma (n=13) with primary glioblastoma (n=13), we found 7 miRNAs to be differentially regulated significantly at an FDR of 0.0% (Figure 1d and Supplementary Table S7).
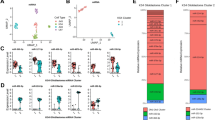
A 23-miRNA Expression Signature Differentiates Glioblastoma from Anaplastic Astrocytoma Accurately
To determine the minimal number of miRNAs that can be used to identify and discriminate glioblastoma and anaplastic astrocytoma cases, we used PAM.14 A 23-miRNA expression signature, identified by PAM, was found to be most discriminatory between glioblastoma and anaplastic astrocytoma (Figure 2a and Table 1). Upon PCA of the 23-miRNA expression signature, most samples that belong to anaplastic astrocytoma (orange triangles) and glioblastoma (blue and purple triangles) formed two distinct groups (Figure 2b). However, there were few samples that were discordant from the original histopathological grading (Figure 2b, see the area under the intersection of the two circles). Prediction accuracy estimation by 10-fold cross-validation using PAM (Figure 3) showed that among 13 anaplastic astrocytoma samples, 11 samples were predicted as anaplastic astrocytoma (cross-validated probability more than 0.5), whereas two samples (CN50 and CN51) were predicted as glioblastoma with an error rate of 0.15. Similarly, among 26 glioblastoma samples used, PAM predicted 24 samples correctly as glioblastoma whereas two samples (CN31 and CN36) as anaplastic astrocytoma with an error rate of 0.078. Thus the 23-miRNA expression signature can discriminate glioblastomas from anaplastic astrocytomas with an overall diagnostic accuracy of 89.7% (Figure 3 and Table 2). The sensitivity for anaplastic astrocytoma was 84.6%, whereas for glioblastoma, it was 92.3%; the specificity for anaplastic astrocytoma was 92.3%, whereas for glioblastoma, it was 84.6% (Table 2).
Details of the PAM-identified 23-miRNA expression signature. (a) One-way hierarchical clustering of PAM identified 23 miRNAs that discriminated glioblastoma from anaplastic astrocytoma. A dual-color code was used, with red and green indicating up- and downregulation, respectively. (b) PCA was performed using the expression values of PAM-identified 23 miRNAs as the most discriminatory between glioblastoma and anaplastic astrocytoma as shown in Table 1. A scatter plot is generated using the first three principal components for each sample (13 anaplastic astrocytoma and 26 glioblastoma tumor samples). The color of the samples is as indicated.
The detailed cross-validated probabilities for anaplastic astrocytoma and glioblastoma tumor samples. The detailed probabilities of 10-fold cross-validation for the 39 malignant astrocytoma samples on the basis of the expression values of the PAM-identified 24 miRNAs are shown. For each sample, its probability as anaplastic astrocytoma (orange color) and glioblastoma (blue color) are shown and it was assigned by the PAM program as either anaplastic astrocytoma or glioblastoma on the basis of which grade's probability is higher. The original histological grade of the samples is shown on the top. The threshold used for training is 2.3. For color figure please see the full text (online) version.
The four samples, which were classified to different grades by PAM, belonged to the progressive pathway (CN50 and CN51=anaplastic astrocytoma; CN31 and CN36=secondary glioblastoma). In view of inherent sampling problems (mainly due to tumor location in the brain) and inadequate sample size available for histological evaluation, there exists a very high possibility of error in grading. Hence, we re-examined the histology of these four discordant cases after processing any additional material that we had in the laboratory. Interestingly, we found that among two secondary glioblastoma samples, one (CN31) showed focal areas of oligodendroglial component and hence this tumor was reclassified as a mixed anaplastic oligoastrocytoma (WHO grade III). Similarly, among two anaplastic astrocytoma samples, histology review in one sample (CN50) after processing additional tumor bits showed the presence of focal areas of necrosis and hence was reclassified as secondary glioblastoma. Thus out of four samples in question as per miRNA expression signature, two samples were actually misclassified originally thereby increasing the overall diagnostic accuracy to 94.8% (Table 2). The sensitivity for anaplastic astrocytoma was 92.3%, whereas for glioblastoma, it was 96.2%; the specificity for anaplastic astrocytoma was 96.2%, whereas for glioblastoma, it was 92.3% (Table 2). It would be interesting to monitor the treatment response and survival of the two patients who remain discordant (CN51 and CN31), which might throw light on the advantage of molecular classification. However, it is worth mentioning that the 23-miRNA expression signature identified all primary glioblastoma samples correctly thus with an accuracy of 100% for primary glioblastoma diagnosis.
Real-Time RT-PCR Validation of Deregulated miRNAs in Selected Malignant Astrocytoma
We performed real-time RT-PCR analysis to validate the differential expression pattern of nine selected miRNAs. These miRNAs were selected on the basis of our microarray data as well as published results. Upon validation of two glioblastoma downregulated miRNAs (miR-219-5p and miR-218), we found that miR-219-5p was specifically downregulated in glioblastoma as compared with anaplastic astrocytomas and normal samples with a P-value of 0.0006 (Figure 4b and Table 3), whereas miR-218 was found to be downregulated in glioblastomas and anaplastic astrocytomas as compared with normal samples with a P-value of 0.009 (Figure 4a and Table 3). Among upregulated miRNAs, we found that miR-21 was upregulated significantly in malignant astrocytomas, particularly in glioblastomas with a P-value of <0.0001 (Figure 4d and Table 3). Three miRNAs (miR-23a, miR-16 and miR-155) were found to be upregulated significantly in particular in glioblastomas with a P-value of 0.0023, 0.0075 and <0.0001, respectively (Figure 4e, f and c, respectively, and Table 3). Three more miRNAs validated in this study are miR-25, miR-93 and miR106b, which are located in the intron (between exon-13 and 14) of the MCM7 gene located in a frequently amplified region (7q21.3–7q22.1) in GBMs (TCGA, 2008). Our validation showed that only miR-25 and miR-93, but not miR-106b, were found upregulated significantly in malignant astrocytomas, particularly glioblastoma with a P-value of 0.0047, 0.0385 and 0.0948, respectively (Figure 4g–i and Table 3). However, it is worth mentioning that whereas miR-106b lacked significance overall, individual comparison showed that it is indeed upregulated in glioblastomas (P=0.0281) and in anaplastic astrocytomas (P=0.0438) as compared with normal samples (Table 3).
Scatter plots of differentially regulated miRNAs in malignant astroctyoma. Log2-transformed miRNA expression ratios obtained from real-time RT-PCR analysis are plotted for miR-218 (a), miR-219-5p (b), miR-155 (c), miR-21 (d), miR-23a (e), miR-16 (f), miR-25 (g), miR-93 (h) and miR-106b (i). Each triangle represents a data derived from one sample. For each sample, fold change in miRNA expression is calculated over its mean expression in normal brain samples.
We also measured the level of these miRNAs in eight different glioma-derived cell lines. We found that miR-218, miR-219-5p, miR-21, miR-23a and miR-155 showed similar expression pattern in more than 75% cell lines tested (Supplementary Figure S3A, B and D). Three miRNAs, miR-25, miR-93 and miR-106b, showed no unique pattern of regulation in the cell lines tested (Supplementary Figure S3C). However, their expression pattern among all cell lines was found to be similar perhaps indicating a coordinated expression from the same transcript.17 Interestingly, miR-16 was found to be downregulated in all glioma cell lines tested unlike its expression in tumor samples (Supplementary Figure S3E).
Modulation of Deregulated miRNAs in Malignant Astrocytoma Affects Transformed Phenotype
To study the biological significance of differentially regulated miRNAs in glioma development, we selected four miRNAs (two upregulated and two downregulated) on the basis of the validation studies described above and potential targets as identified by bioinformatics analysis (data not shown). For the two upregulated miRNAs, miR-21 and miR-23a, we used a loss-of-function approach in glioma cell lines and monitored the effect on cell proliferation, chemosensitivity and soft agar colony formation. Transfection of U138 glioma cells with anti-miR inhibitors specific to miR-21 and miR-23a resulted in substantial reduction in their levels up to 72 h after transfection in comparison with Cy3 dye-labeled anti-miR-negative control, which is a non-targeting random nucleotide sequence (Figure 5a and b). Whereas downregulation of miR-21 and miR-23a did not have any significant effect on the proliferative capacity of U138 cells (data not shown), it resulted in significant reduction in soft agar colony formation capacity (Figure 5c). Furthermore, downregulation of miR-21 rendered cells significantly sensitive to the chemotherapeutic drug etoposide (Figure 5d and e).
Effect of downregulation of miR-21 and miR-23a and overexpression of miR-218 and miR-219-5p in U138 glioma cells. (a, b) U138 cells were transfected with either a Cy3 dye-labeled anti-miR-negative control or with anti-miR inhibitors specific to miR-21 and miR-23a. At the indicated time points after transfection, the levels of mature miR-21 (a) or miR-23a (b) were measured as a log2 ratio and plotted. Fold change in miRNA expression in specific anti-miR-transfected cells is calculated over its expression in control anti-miR-transfected cells. (c) U138 cells were transfected with either the Cy3 dye-labeled anti-miR-negative control or with anti-miR inhibitors specific to miR-21 and miR-23a. The cells were harvested after 36 h of transfection and 5 × 103 cells, in duplicates, were plated for soft colony formation as described under Materials and methods. The number of colonies were counted after 2 weeks and plotted with the total number of colonies in the control sample considered as 100%. (d) U138 cells were transfected with either the Cy3 dye-labeled anti-miR-negative control or with an miR-21-specific anti-miR inhibitor. After 36 h of transfection, the cells (3 × 103) were treated with different concentrations of etoposide, as indicated, in triplicates. After 48 h of drug treatment, the proportion of live cells was quantified by MTT assay. The absorbance of control cells was considered as 100%. (e) The proportion of viable cells at concentration of 4, 6 and 8 μg/ml as measured in panel d is shown. The difference in viability between control and anti-miR-21-treated samples at 4 and 6 μg/ml was found to be significant as analyzed by Student's t-test (P-value=0.006 and 0.05, respectively). (f, g) U138 cells were transfected with either Cy3 dye-labeled Pre-miR-negative control or Pre-miR precursors specific to miR-218 and miR-219-5p. At the indicated time points after transfection, the levels of mature miR-218 (f) and miR-219 (g) were measured as a log2 ratio and plotted. Fold change in miRNA expression in specific Pre-miR-transfected cells is calculated over its expression in control Pre-miR-transfected cells. (h) U138 cells were transfected with either Cy3 dye-labeled Pre-miR-negative control or Pre-miR precursors specific to miR-218 and miR-219-5p in 24 multi-well plates. At the indicated time points, the cells were harvested and viable cells were counted and plotted. (i) U138 cells were transfected with either Cy3 dye-labeled Pre-miR-negative control or Pre-miR precursors specific to miR-218 and miR-219-5p. The cells were harvested after 36 h of transfection and 5 × 103 cells, in duplicates, were plated for soft colony formation as described under Materials and methods. The number of colonies were counted after 2 weeks and plotted with the total number of colonies in the control sample as 100%.
For two downregulated miRNAs, miR-218 and miR-219-5p, we overexpressed by transfecting the Pre-miR precursor molecules into glioma cells and monitored the effect on cell proliferation, chemosensitivity and soft agar colony formation. Transfection of U138 glioma cells with Pre-miR precursors specific to miR-218 and miR-219-5p resulted in very high elevated levels of mature species of respective miRNAs up to 72 h after transfection in comparison with the Cy3 dye-labeled Pre-miR-negative precursor, which is a non-targeting random sequence molecule (Figure 5f and g). While over expression of miR-218 resulted in substantial reduction in cell proliferation, miR-219-5p inhibited proliferation marginally (Figure 5h). However, upon overexpression, both miRNAs inhibited soft agar colony formation significantly (Figure 5i). Thus these results put together suggest that these two sets of miRNAs (miR-21 and miR-23a; miR-218 and miR-219-5p) may have opposing roles in regulating tumorigenesis.
Discussion
Treatment options for patients with malignant glioma often depend on the histological grade of the tumor. The standard therapeutic protocol for grade IV glioblastoma patients includes surgery followed by radiotherapy with concomitant, and further, cyclical chemotherapy (temozolamide).6 Some studies showed a small, but significant improvement in survival of grade III patients with the use of chemotherapy in addition to irradiation.18, 19 However, chemotherapy failed to consistently improve the outcome of patients with anaplastic astrocytoma.7 Furthermore, the length of survival is distinctly different between anaplastic astrocytoma and glioblastoma patients.8 As the currently used histology-based grading is subjective, there is a need for more accurate methods of classification. Adding more complexity, glioblastomas are further divided into primary glioblastoma or secondary glioblastoma on the basis of their clinical presentations. However, they have an equally poor prognosis when adjusted for patient age.11
Several groups have used global transcript profiling to obtain gene expression signatures that reflected the biology of different classes of astrocytoma and also provided markers for use in diagnosis and clinical management.11, 20, 21 Copy number variation studies using genome-wide array-CGH profiles have identified many regions of amplification and deletions harboring many genes found to be important in glioma development.22, 23, 24 However, these findings and related signatures are yet to be translated to utility in clinical settings suggesting that these studies require further characterization and validation.
There have been many studies reporting the role of specific miRNAs in glioma development.25 Among them, specific studies describe the target identification and functional characterization of the following miRNAs: miR-34a, miR-128, miR-7, miR-21 and miR-26a.26, 27, 28, 29, 30, 31, 32, 33 There are also studies of expression profiling analyzing the expression of large number of miRNAs in malignant glioma.25, 27, 34, 35, 36 However these studies profiled fewer number of miRNA and used few number of glioma samples.
In this study, we have profiled the expression of 756 miRNAs using a sample set of 39 malignant astrocytoma samples. Analysis of microarray data identified a total of 84 miRNAs that are differentially regulated in malignant astrocytoma in comparison with normal brain thus bringing the total proportion of deregulated miRNAs to 11.11%. Using a subset of SAM-identified significant miRNAs, PCA was able to group normal, grade III and grade IV into three distinct clusters indicating the power and utility of miRNA gene expression signature for tumor classification and grading.
We also used SAM to identify miRNA signatures that can differentiate secondary glioblastoma and primary glioblastoma from anaplastic astrocytoma, secondary glioblastoma from primary glioblastoma as well as progressive astrocytoma from de novo glioblastoma. Of these, particularly interesting was the comparison between secondary glioblastoma and primary glioblastoma with anaplastic astrocytoma, which yielded miRNAs, which are common (n=50) as well as unique to secondary glioblastoma (n=18) and primary glioblastoma (n=26). These two classes of miRNAs may reflect a large similarity between secondary glioblastoma and primary glioblastoma, and their uniqueness as they arise through different pathways. Additionally, comparison of progressive astrocytoma samples (anaplastic astrocytoma and secondary glioblastoma) with primary glioblastoma yielded 21 differentially regulated miRNAs. Surprisingly, a direct comparison of secondary glioblastoma with primary glioblastoma yielded only seven differentially regulated miRNA perhaps suggesting an overall similarity between secondary glioblastoma and primary glioblastoma. However, the miRNA signatures identified in this study as unique to a specific grade or pathway may characterize specific biological pathways responsible for the development of a progressive vs de novo pathway of astrocytoma.
Furthermore, we identified a 23-miRNA expression signature using PAM, which could discriminate glioblastoma from anaplastic astrocytoma with an overall accuracy of 94.8%. Interestingly, the misclassified samples belonged to the progressive astrocytoma group. The progression may often make it difficult to grade precisely these samples because of the presence of grade-specific histological features in different areas of tumor tissue. In contrast to this, all the primary glioblastoma cases were accurately identified. It is interesting to note that out of 23 miRNAs identified by PAM, 21 were upregulated in glioblastoma in comparison with anaplastic astrocytoma whereas two were downregulated in glioblastoma in comparison with anaplastic astrocytoma.
Furthermore, we have confirmed the grade-specific regulation of nine selected miRNAs by RT-qPCR on an independent set of malignant astrocytoma samples. Corroboratively, eight of these miRNAs (except miR-16) showed similar expression pattern in the majority of the glioma-derived cell lines. For miR-21 and miR-23a, while the specific anti-miR-mediated downregulation in glioma cells did not show any change in cell proliferative capacity, it resulted in significant reduction in their ability to form colonies in soft agar, suggesting that these miRNAs might have an important role in astrocyte transformation. Furthermore, downregulation of miR-21 increased the chemosensitivity of glioma cells to etoposide. These results suggest that miR-21 and miR-23a may target some proapoptotic or growth-inhibitory genes thus contributing to tumorigenesis. Indeed, miR-21 has been shown to target the components of the p53 and TGF-β pathway, mitochondrial apoptosis-related genes as well as RECK and TIMP3 (tissue inhibitors of matrix metalloproteinases) thereby contributing to tumorigenicity and invasion.30, 32 For miR-218 and miR-219-5p, exogenous overexpression by transfection of the specific Pre-miRs reduced the cell proliferation, with miR-218 being highly growth-inhibitory. Furthermore, we found both that miRNAs inhibited soft agar colony formation. It has been shown that overexpression of zebrafish miR-219 leads to induction of apoptosis during zebrafish development.37 On target prediction analysis, we found that miR-219-5p may target EGFR1, a gene very frequently amplified in primary glioblastoma (data not shown). miR-218 has been found to be downregulated in cervical cancer in an E6/E7-dependent manner and consequent upregulation of the miR-218 target LAMB3 contributing to tumorigenicity.38 These results together suggest that miR-218 and miR-219-5p may target some oncogenic or antiapoptotic molecules and may have an important role in astrocytoma development.
Our study with a list of deregulated miRNAs in malignant astrocytoma would also form a basis for further investigation of the identification of their targets, thus leading to a better understanding of glioma development and progression. Furthermore, precise identification of grade IV is essential for choosing the correct treatment option and clinical management. Although histology is still the gold standard for grading astrocytomas, there exists the possibility of inaccurate grading, which necessitates a more robust molecular classifier. Our finding that the 23-miRNA expression signature could accurately (94.8%) discriminate glioblastoma from anaplastic astrocytoma tumors raises the possibility of using such discriminatory miRNAs to develop a rapid and accurate molecular diagnostic test of glioblastoma vs anaplastic astrocytoma in the future. Indeed, compared with the mRNA profiling that has been proposed for diagnosis of various cancers, miRNA profiling appears to have several advantages.39, 40 Nonetheless, further validation of these discriminatory miRNAs in a large number of patients and in independent studies is necessary before clinical application becomes realistic.
References
Yue J, Tigyi G . MicroRNA trafficking and human cancer. Cancer Biol Ther 2006;5:573–578.
Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature 2005;435:834–838.
Calin GA, Sevignani C, Dumitru CD, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA 2004;101:2999–3004.
Landis SH, Murray T, Bolden S, et al. Cancer statistics, 1999. CA Cancer J Clin 1999;49:8–31.
Liau LM, Becker DP, Cloughesy TF, et al. (eds). Brain Tumor Immunotherapy. Humana Press: Totowa, 2001, pp 3.
Reardon DA, Wen PY . Therapeutic advances in the treatment of glioblastoma: rationale and potential role of targeted agents. Oncologist 2006;11:152–164.
Stupp R, Reni M, Gatta G, et al. Anaplastic astrocytoma in adults. Crit Rev Oncol Hematol 2007;63:72–80.
Legler JM, Ries LA, Smith MA, et al. Cancer surveillance series [corrected]: brain and other central nervous system cancers: recent trends in incidence and mortality. J Natl Cancer Inst 1999;91:1382–1390.
Barnett GH, (ed). High-Grade Gliomas: Diagnosis and Treatment. Humana Press: Totowa, 2007, pp 3.
Ohgaki H, Kleihues P . Genetic pathways to primary and secondary glioblastoma. Am J Pathol 2007;170:1445–1453.
Furnari FB, Fenton T, Bachoo RM, et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev 2007;21:2683–2710.
Somasundaram K, Reddy SP, Vinnakota K, et al. Upregulation of ASCL1 and inhibition of Notch signaling pathway characterize progressive astrocytoma. Oncogene 2005;24:7073–7083.
Ritchie ME, Silver J, Oshlack A, et al. A comparison of background correction methods for two-colour microarrays. Bioinformatics 2007;23:2700–2707.
Tibshirani R, Hastie T, Narasimhan B, et al. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci USA 2002;99:6567–6572.
Das S, El-Deiry WS, Somasundaram K . Regulation of the p53 homolog p73 by adenoviral oncogene E1A. J Biol Chem 2003;278:18313–18320.
Tusher VG, Tibshirani R, Chu G . Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 2001;98:5116–5121.
Petrocca F, Vecchione A, Croce CM . Emerging role of miR-106b-25/miR-17-92 clusters in the control of transforming growth factor beta signaling. Cancer Res 2008;68:8191–8194.
Walker MD, Green SB, Byar DP, et al. Randomized comparisons of radiotherapy and nitrosoureas for the treatment of malignant glioma after surgery. N Engl J Med 1980;303:1323–1329.
Stewart LA . Chemotherapy in adult high-grade glioma: a systematic review and meta-analysis of individual patient data from 12 randomised trials. Lancet 2002;359:1011–1018.
Reddy SP, Britto R, Vinnakota K, et al. Novel glioblastoma markers with diagnostic and prognostic value identified through transcriptome analysis. Clin Cancer Res 2008;14:2978–2987.
Liang Y, Diehn M, Watson N, et al. Gene expression profiling reveals molecularly and clinically distinct subtypes of glioblastoma multiforme. Proc Natl Acad Sci USA 2005;102:5814–5819.
Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 2008;455:1061–1068.
Wiedemeyer R, Brennan C, Heffernan TP, et al. Feedback circuit among INK4 tumor suppressors constrains human glioblastoma development. Cancer Cell 2008;13:355–364.
Beroukhim R, Getz G, Nghiemphu L, et al. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proc Natl Acad Sci USA 2007;104:20007–20012.
Silber J, Lim DA, Petritsch C, et al. miR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Med 2008;6:14.
Li Y, Guessous F, Zhang Y, et al. MicroRNA-34a inhibits glioblastoma growth by targeting multiple oncogenes. Cancer Res 2009;69:7569–7576.
Godlewski J, Nowicki MO, Bronisz A, et al. Targeting of the Bmi-1 oncogene/stem cell renewal factor by microRNA-128 inhibits glioma proliferation and self-renewal. Cancer Res 2008;68:9125–9130.
Zhang Y, Chao T, Li R, et al. MicroRNA-128 inhibits glioma cells proliferation by targeting transcription factor E2F3a. J Mol Med 2009;87:43–51.
Kefas B, Godlewski J, Comeau L, et al. MicroRNA-7 inhibits the epidermal growth factor receptor and the Akt pathway and is downregulated in glioblastoma. Cancer Res 2008;68:3566–3572.
Papagiannakopoulos T, Shapiro A, Kosik KS . MicroRNA-21 targets a network of key tumor-suppressive pathways in glioblastoma cells. Cancer Res 2008;68:8164–8172.
Chen Y, Liu W, Chao T, et al. MicroRNA-21 downregulates the expression of tumor suppressor PDCD4 in human glioblastoma cell T98G. Cancer Lett 2008;272:197–205.
Gabriely G, Wurdinger T, Kesari S, et al. MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Mol Cell Biol 2008;28:5369–5380.
Huse JT, Brennan C, Hambardzumyan D, et al. The PTEN-regulating microRNA miR-26a is amplified in high-grade glioma and facilitates gliomagenesis in vivo. Genes Dev 2009;23:1327–1337.
Ciafre SA, Galardi S, Mangiola A, et al. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem Biophys Res Commun 2005;334:1351–1358.
Chan JA, Krichevsky AM, Kosik KS . MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res 2005;65:6029–6033.
Malzkorn B, Wolter M, Liesenberg F, et al. Identification and functional characterization of microRNAs involved in the malignant progression of gliomas. Brain Pathol 2009;20:539–550.
Zhang MC, Lv Y, Qi YT, et al. [Knockdown and overexpression of miR-219 lead to embryonic defects in zebrafish development]. Fen Zi Xi Bao Sheng Wu Xue Bao 2008;41:341–348.
Martinez I, Gardiner AS, Board KF, et al. Human papillomavirus type 16 reduces the expression of microRNA-218 in cervical carcinoma cells. Oncogene 2008;27:2575–2582.
Bertucci F, Viens P, Tagett R, et al. DNA arrays in clinical oncology: promises and challenges. Lab Invest 2003;83:305–316.
Sotiriou C, Piccart MJ . Taking gene-expression profiling to the clinic: when will molecular signatures become relevant to patient care? Nat Rev Cancer 2007;7:545–553.
Acknowledgements
This study was supported by a grant from DBT, Government of India. KS is a Wellcome Trust International Senior Research Fellow. Infrastructural support by funding from ICMR, DBT, DST and UGC to MCB is acknowledged. We thank Subhomoy Mukherjee, Lyndsay Priscilla A and T Ganesh Babu for technical help. AMS gratefully acknowledges the JRF from CSIR. The authors acknowledge Profs. MRS Rao, P Kondaiah, AS Hegde, BA Chandramouli, A Arivazhagan and A Balasubramanian for their help.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Modern Pathology website
Supplementary information
Rights and permissions
About this article
Cite this article
Rao, S., Santosh, V. & Somasundaram, K. Genome-wide expression profiling identifies deregulated miRNAs in malignant astrocytoma. Mod Pathol 23, 1404–1417 (2010). https://doi.org/10.1038/modpathol.2010.135
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2010.135
Keywords
This article is cited by
-
The Role of microRNA-23a-3p in the Progression of Human Aging Process by Targeting FOXO3a
Molecular Biotechnology (2024)
-
MicroRNA-770 affects proliferation and cell cycle transition by directly targeting CDK8 in glioma
Cancer Cell International (2018)
-
Overexpression of miR-21 in stem cells improves ovarian structure and function in rats with chemotherapy-induced ovarian damage by targeting PDCD4 and PTEN to inhibit granulosa cell apoptosis
Stem Cell Research & Therapy (2017)
-
MicroRNA profiling of low-grade glial and glioneuronal tumors shows an independent role for cluster 14q32.31 member miR-487b
Modern Pathology (2017)
-
Expression of microRNAs in tumors of the central nervous system in pediatric patients in México
Child's Nervous System (2017)