Abstract
Glial fibrillary acidic protein (GFAP) is a member of the intermediary filament protein family. It is an important component of astrocytes and a known diagnostic marker of glial differentiation. GFAP is expressed in other neural tumors and pleomorphic adenoma and, less frequently, in cartilage tumors, chordomas, and soft tissue myoepitheliomas. The aim of this study was to evaluate the role of GFAP and its reliability in nonglial tumors as an immunohistochemical marker. We evaluated GFAP gene and protein expression using Q-PCR and immunohistochemistry, respectively, in 81 and 387 cases of soft tissue, bone tumors, and salivary pleomorphic adenomas. Immunohistochemistry staining for GFAP was observed in all osteosarcomas (8 cases), all pleomorphic adenomas (7 cases), in 5 of 6 soft tissue myoepitheliomas, and in 21 of 76 chondrosarcomas. By Q-PCR, GFAP was highly expressed in pleomorphic adenomas and, to a lesser extent, chondrosarcomas, soft tissue myoepitheliomas, and chondroblastic osteosarcomas. The results that we obtained by immunohistochemistry and Q-PCR were well correlated. GFAP is a potential marker for tumors with cartilaginous differentiation, supported by evidence that GFAP is expressed in certain cases of myoepithelial tumors by immunohistochemistry, including soft tissue myoepitheliomas, which are related to cartilaginous differentiation. These findings contribute significantly to the diagnosis of soft tissue myoepitheliomas with cartilaginous differentiation and chondroblastic osteosarcoma in mesenchymal tumors.
Similar content being viewed by others
Main
Intermediate filaments are proteins that are associated with plasma membranes and have structural function, reinforcing, and organizing cells in tissues. Their most important function is to provide mechanical support for the plasma membrane, which is responsible for contact with cells or with the extracellular matrix.
Intermediate filaments are grouped into six major classes: type I (acidic keratins), type II (basic keratins), type III (mesenchymal tissues intermediate filaments), type IV (neurofilaments), nonstandard type IV, and type V (laminins). The type III intermediate filament group comprises vimentin, desmin, peripherin, and glial fibrillary acid protein and is found in many tissues. One of the notable molecular aspects of the type III intermediate filament is its capacity to form hetero- and homopolymeric intermediate filaments.1
Glial fibrillary acidic protein (GFAP) is a type III intermediate filament. It is a 51-kD protein and has the smallest HEAD domain among the type III intermediate filament proteins. As do other type III intermediate filaments, the assembly of GFAP is regulated by phosphorylation–dephosphorylation of the HEAD domain, wherein its charge is altered. This assembly contributes to extensive remodeling of glial frameworks during mitosis.
Another type III intermediate filament protein, vimentin, colocalizes with GFAP in immature, reactive, and radial glia, suggesting that vimentin regulates glial architecture.1
GFAP is the major component of several cell types of neuroglia, such as astrocytes, ependymal cells, and Muller cells of the retina. In the central nervous system, it is expressed in ependymal cells and choroid plexus epithelium but is absent from mature oligodendrogliocytes.2 Among nonglial tissues, GFAP is expressed in Schwann cells, Küpffer cells of the liver, and interstitial cells of the pituitary gland and paraganglions.2 In addition, GFAP is expressed in osteocytes of mature bones and chondrocytes from epiglottal, bronchial, and tracheal cartilage.2, 3, 4, 5, 6
GFAP is the most specific marker for cells of astrocytic origin under normal and pathological conditions. By immunohistochemistry, however, GFAP has been shown to be expressed in mesenchymal tumors, such as soft tissue myoepitheliomas, peripheral nerve sheath tumors, and chordomas,3, 7, 8, 9, 10, 11, 12 and also in mixed tumors, such as salivary gland pleomorphic adenomas.13, 14 These findings can create problems for the diagnosis of tumors that have similar morphologies and are positive for GFAP, such as myoepithelioma and chondroblastic osteosarcoma of the head and neck. However, GFAP expression can be useful in the differential diagnosis of mesenchymal tumors, especially for small biopsies.
In this study, we evaluated GFAP expression using real-time PCR and immunohistochemistry in mesenchymal tumors and salivary pleomorphic adenomas.
Materials and methods
Samples
Eighty-one tumors from the tissue biorepository of Hospital A. C. Camargo, Sao Paulo, Brazil, were used for Q-PCR experiments. A detailed description of the samples is presented in Table 1.
All samples were collected during surgical procedures from 1997–2007. Tissue samples that were obtained from surgeries were snap-frozen in liquid nitrogen, and biopsy samples were collected in RNAlater- (Ambion®, Austin, TX, USA). All samples were stored at −140°C until further processing. Histopathological diagnoses were reviewed by two pathologists (IWC, RF) before RNA extraction. Frozen samples were hand dissected to remove necrotic tissue and to enrich the tumor. Total RNA was extracted using Trizol- (Life Technologies, Inc., Grand Island, NY, USA).
For the immunohistochemical study, 387 formalin-fixed, paraffin-embedded tumors were obtained from the anatomical pathology files of Hospital A. C. Camargo. All cases were reviewed by two pathologists (IWC, RF), and immunohistochemistry was performed using a panel of antibodies for proper tumor classification.
A detailed description of the samples is presented in Table 1. The analyzed samples were spotted onto tissue microarray slides, as described.15
Immunohistochemistry
Tissue sections were mounted on silane-coated glass slides (3-aminopropyltriethoxysilane) and dried for 30 min at 37°C. The sections were deparaffinized in xylene and rehydrated through a series of graded alcohols. Endogenous peroxidase activity was blocked by incubating the sections in a methanol bath that contained 3% hydrogen peroxide for 20 min, which was followed by washes in distilled water. All sections were initially submitted to heat-induced epitope retrieval using citrate buffer (pH 6.0).
The primary GFAP antibody (polyclonal, DAKO®, CA, USA, working dilution 1:10 000) was added and incubated for 90 min at room temperature. Preliminary testing was performed in our laboratory to determine the optimal concentration for the primary antibody and to select negative and positive controls using the dilution data that were supplied by the manufacturer. After washing the primary antibody away with phosphate-buffered saline, we add the polymer advance (DAKO) for 20 min.
Freshly prepared diaminobenzidine solution (one drop of 3,30-diaminobenzidine tetrahydrochloride per 1 ml of substrate, DAKO-) was added to each section for 2 min. Diaminobenzidine solution was removed by rinsing with distilled water. The slides were counterstained with hematoxylin, dehydrated in ethanol, cleared in xylene, and mounted using Entelan.16
Evaluation of Immunohistochemistry
Staining was evaluated independently by two observers (GCS and IWC). The intensity of staining reactions was divided into three groups with regard to cytoplasmic staining: 0, negative; 1, weak positivity; 2, strong positivity.
Q-Pcr
Aliquots of 2 μg of total RNA were reverse transcribed using the High-Capacity cDNA Reverse Transcription kit (Applied Biosystems, CA, USA), following the manufacturer's instructions. Primer pairs for real-time PCR (forward CTGGAGGTTGAGAGGGACAA and reverse CTCCAGCGACTCAATCTTCC) were selected with Primer 3.0 (http://frodo.wi.mit.edu). Reactions were performed with 10 ng cDNA, using the SYBR Green Master Mix system according to the manufacturer's instructions.
Reactions were run in duplicate on an ABI PRISM- 7300 Sequence Detection System (Applied Biosystems) and analyzed using Sequence Detection Software (version 2.3). The Ct values were transformed to quantities using the comparative Ct method,17 and normalization factors for each sample were calculated using Genorm software, based on the expression levels of GAPDH, β-actin, and HPRT genes, as described by Vandesompele et al.18 As a reference, we used a pool of RNA that was obtained from 15 distinct human cell lines, and relative expression values were expressed on a logarithmic scale (log 2). One astrocytoma sample was used as a reference for positive GFAP expression in our analysis.
Statistical Analysis
SPSS, version 12.0 was used for statistical analysis; t test was used to compare two groups of numerical variables, and ANOVA was used for more than two groups. The Spearman coefficient was used to evaluate the correlation between groups; χ2 was used to evaluate the difference in frequencies between groups.
Results
Immunohistochemistry
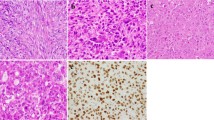
The results of GFAP immunostaining in different histological tumor types are compiled in Table 2; examples are shown in Figure 1. GFAP expression was observed in 100 (26%) cases. Most of the positive cases showed weak positive immunostaining, and 13 (3%) cases expressed GFAP robustly. Strong positive staining was observed in pleomorphic adenomas (5/7), soft tissue myoepitheliomas (2/6), chondrosarcomas (4/76), and chondroblastic osteosarcomas (2/3) (Figure 1).
Mesenchymal tumors: immunostaining for GFAP. (a) Chondroblastic osteosarcoma (strong positivity, × 100). (b) Soft tissue myoepitelioma (strong positivity, × 100). (c) Chondrosarcoma (strong positivity, × 200). (d) Chondrosarcoma (negative, × 400). (e) Liposarcoma (weak positivity, × 400). (f) Synovial sarcoma (negative, × 100).
For statistical analysis of the immunohistochemistry staining, we separated the tumor types in two groups with regard for their potential for chondroblastic differentiation.
GFAP expression in the tumors with potential cartilaginous differentiation (pleomorphic adenomas, chondroblastic osteosarcomas, soft tissue myoepitheliomas, and chondrosarcomas) was significantly higher compared with the group without it (P<0.001). Additionally, there were no other mesenchymal tumors that expressed GFAP strongly.
Q-PCR
Overall, tumors that underwent cartilaginous differentiation had higher GFAP expression levels (Figure 2). Considering the histological types of individual tumors, we observed that pleomorphic adenomas (n=7) upregulated GFAP, as did soft tissue myoepiteliomas (n=3) and chondrosarcomas (n=4). There was no significant difference in expression between osteoblastic osteosarcomas (n=8) and fibroblastic tumors (n=7), pleomorphic sarcomas (n=6), leiomyosarcoma (n=6), or synovial sarcomas (n=10).
Expression of GFAP evaluated by Q-PCR in pleomorphic adenomas (PA), chondrosarcomas (CD), soft tissue myoepiteliomas (ME), chondroblastic osteosarcomas (OS w/C), osteoblastic osteosarcomas (OS no/C), liposarcomas (LPS), pleomorphic sarcomas (PS), fibroblastic tumors (FT), synovial sarcomas (SS), and muscular tumors (MT). Q-PCR was performed using SYBR green.
We observed a significant difference between tumors that had the potential for cartilaginous differentiation [pleomorphic adenomas (n=7), soft tissue myoepiteliomas (n=3), chondrosarcomas (n=4), and chondroblastic osteosarcomas (n=4)] and tumors that did not [osteoblastic ostesarcomas (n=8), synovial sarcomas (n=10), liposarcomas (n=3), pleomorphic sarcomas (n=6), fibroblastic tumors (n=7), and muscular tumors (n=9), including alveolar soft part sarcomas] (P<0.0001, Figure 3). GFAP mRNA expression was undetected in 17 sarcoma samples (4 synovial sarcomas, 4 pleomorphic sarcomas, 4 leiomyosarcomas, 3 desmoid-type fibromatosis, and, 2 fibrosarcomas). These cases were excluded from statistical analysis.
Expression of GFAP, as evaluated by Q-PCR, comparing all tumors that showed cartilaginous differentiation (pleomorphic adenomas, soft tissue myoepiteliomas, chondrosarcomas, and chondroblastic osteosarcomas) with tumors without cartilaginous differentiation (osteoblastic ostesarcomas, synovial sarcomas, liposarcomas, pleomorphic sarcomas, fibroblastic tumors, and muscular tumors).
For 40 cases, we performed both immunohistochemistry and RT–PCR, and the concordance between immunohistochemistry and RT–PCR expression was highly significant (P<0.001), as shown in Figure 4.
Discussion
GFAP expression is the most reliable marker of glial differentiation. We observed an increase in both GFAP mRNA and protein levels in mesenchymal tumors and salivary gland pleomorphic adenomas, most of which presented with cartilaginous differentiation (pleomorphic adenomas, chondrosarcomas, soft tissue myoepitheliomas, and osteosarcomas). Our results are strong evidence that as an intermediate filament, GFAP regulates cartilaginous differentiation.
Expression of GFAP in nonglial cells has been described in mature osteocytes and chondrocytes (13/20 osteocyte samples from the vertebral body, 5/11 chondrocyte samples from the epiglottis, 3/11 chondrocyte samples from rib cartilage, 2/6 samples from tracheal cartilage, and 4/5 samples from bronchial cartilage),3 wherein strong cytoplasmatic staining was observed in chondrocytes from bronchial cartilage and tracheal samples stained weakly.5 These findings suggest that osteocytes and chondrocytes have intermediate filaments in their cytoskeleton that are identical to, are similar to, or associate with GFAP.3 Chondrocytes in elastic cartilage, stain positive for GFAP more frequently than those from nonelastic cartilage (fibrous or hyaline),8 as do chondrocytes from the epiglottis. These findings suggest a histogenetic relationship between epiglottal cartilage and the neural crest.4
In our samples, 28% (21/76) of chondrosarcomas expressed GFAP at the protein level. Therefore, the majority of chondrosarcomas failed to express GFAP, which can be explained by the observations that immature cartilage increases GFAP expression relative to mature cartilage5, 8 and that the number of GFAP-positive chondrocytes decreases from fetal stages to adulthood.6
The etiopathogenesis of chondrosarcomas follows two pathways: originating from a mesenchymal precursor cell or developing from dedifferentiation of mature chondrocytes. Thus, GFAP-positive chondrosarcomas represent unique tumors that originate in immature chondrocytes. Other tumors that experience heterologous cartilage differentiation, such as chondroblastic osteosarcomas, soft tissue myoepitheliomas with chondroblastic differentiation, and pleomorphic adenomas, express GFAP more frequently than chondrossarcomas.
In another study, the authors acquired samples from immature and mature teratomas and classified them into four grades according to their stage of maturity.8 They showed that GFAP-positive chondrocytes were abundant in immature cartilage and that as the cartilage matured, these chondrocytes decreased in number and distributed peripherally. Furthermore, in immature teratomas, GFAP-positive cartilage neared central nervous tissue.8 Taking these data into consideration, we speculate that GFAP is expressed in the early stages of differentiation and that GFAP-positive chondrosarcomas originate in immature cartilaginous cells instead of through dedifferentiation of mature chondrocytes.
Another possible explanation for the absence of GFAP in chondrosarcomas concerns the observation that joint cartilage does not express GFAP.5 These authors reported GFAP immunoexpression in three chondrosarcomas, and in all cases, the expression was restricted to only several cells. They also showed that all joint cartilage cases were negative. We also evaluated all chondrosarcomas by immunohistochemistry with regard to their location (axial or members) and their differentiation (grades 1, 2, or 3), but no differences were found.
Soft tissue myoepitheliomas frequently express GFAP.7, 10, 11, 12 In our study, five of six soft tissue myoepitheliomas were positive for GFAP. Cartilaginous differentiation was present in two cases, which were strongly positive, as evaluated by immunohistochemistry. Other authors have described GFAP positivity in 46% of soft tissue myoepitheliomas (40/87) by immunohistochemistry, wherein 24 samples stained robustly.7 In their study, there were six cartilaginous metaplasia, six tumors that showed osseous differentiation, and four tumors that showed cartilaginous and osseous differentiation. These cases, however, were not evaluated with regard to GFAP expression, and we could not determine whether expression was associated with cartilaginous or osseous differentiation.
GFAP expression also has been reported in an isolated case of gliosarcoma that had characteristics of chondroblastic osteosarcoma, which was confirmed by immunostaining and electronic microscopy.9
In our study, all cases of osteosarcomas were positive for GFAP. The strongest expression was documented in tumors with chondroblastic differentiation. In contrast to cartilaginous cells, expression of GFAP is observed in mature osteocytes from vertebral bodies.3
Our results, obtained by Q-PCR, which also correlate with the immunohistochemical findings, confirm that GFAP is a bona fide marker for cartilaginous differentiation.
We also measured expression patterns in soft tissue tumors without cartilaginous expression by cDNA microarray in 102 soft tissue tumors, including desmoid-type fibromatosis (n=19), fibrosarcomas (n=6), malignant peripheral nerve sheath tumors (n=5), neurofibromas (n=3), schwanomas (n=2), leiomyosarcomas (n=19), leiomyomas (n=2), alveolar soft part sarcomas (n=2), gastrointestinal stromal tumors (n=2), liposarcomas (n=6), synovial sarcomas (n=19), and pleomorphic sarcomas (n=16). [The accession number for the raw data is GSE14541 (http://www.ncbi.nlm.nih.gov/projects/geo)]. Only one case of leiomyosarcoma and one malignant peripheral nerve sheet tumor upregulated GFAP expression (data not shown). We also compared the pattern of expression between malignant and benign tumors but did not find any difference.
This study suggests that GFAP is related to cartilaginous differentiation. GFAP has been used as a marker for soft tissue myoepithelial tumors. This expression, however, might be because of cartilaginous differentiation, not to myoepithelial cells. Therefore, immunohistochemical expression of GFAP should be performed carefully in the diagnosis of mesenchymal tumors, because not only can soft tissue myoepitheliomas express it but other tumors that are capable of cartilaginous and osseous differentiation can as well.
Accession codes
References
Kierszenbaum AL . Basic tissues and integrated cell biology. In: Kierszenbaum AL (ed) Histology and Cell Biology, 2nd edn Mosby Elsevier: Philadelphia, 2007, pp 1–54.
DeLellis RA, Shin SJ . Immunohistology of endocrine tumors. In: Dabbs D (ed) Diagnostic Immunohistochemistry, 2nd edn Churchill Livingstone: Philadelphia, 2006, pp 261–296.
Kasantikul V, Shuangshoti S . Positivity to glial fibrillary acidic protein in bone, cartilage, and chordoma. J Surg Oncol 1989;41:22–26.
Kepes JJ, Perentes E . Glial fibrillary acidic protein in chondrocytes of elastic cartilage in the human epiglottis: an immunohistochemical study with polyvalent and monoclonal antibodies. Anat Rec 1988;220:296–299.
Dolman CL . Glial fibrillary acidic protein and cartilage. Acta Neuropathol (Berl) 1989;79:101–103.
Viale G, Doglioni C, Dell'Orto P, et al. Glial fibrillary acidic protein immunoreactivity in human respiratory tract cartilages and pulmonary chondromatous hamartomas. Am J Pathol 1998;133:363–373.
Hornick JL, Fletcher CD . Myoepithelial tumors of soft tissue: a clinicopathologic and immunohistochemical study of 101 cases with evaluation of prognostic parameters. Am J Surg Pathol 2003;27:1183–1196.
Notohara K, Hsueh CL, Awai M . Glial fibrillary acidic protein immunoreactivity of chondrocytes in immature and mature teratomas. Acta Pathol Jpn 1990;40:335–342.
Hayashi K, Ohara N, Jeon HJ, et al. Gliosarcoma with features of chondroblastic osteosarcoma. Cancer 1993;72:850–855.
Kuhnen C, Herter P, Kasprzynski A, et al. Myoepithelioma of soft tissue—case report with clinicopathologic, ultrastructural and cytogenetic findings. Pathologe 2005;26:331–337.
Kilpatrick SE, Hitchcock MG, Kraus MD, et al. Mixed tumors and myoepitheliomas of soft tissue: a clinicopathologic study of 19 cases with a unifying concept. Am J Surg Pathol 1997;21:13–22.
Neto AG, Pineda-Daboin K, Luna MA . Myoepithelioma of the soft tissue of the head and neck: a case report and review of the literatura. Head Neck 2004;26:470–473.
Shah SS, Chandan VS, Wilbur DC, et al. Glial Fibrillary acidic protein and CD57 immunolocalization in cell block preparations is a useful adjunct in the diagnosis of pleomorphic adenoma. Arch Pathol Lab Med 2007;131:1373–1377.
Curran AE, White DK, Damm DD, et al. Polymorphous low-grade adenocarcinoma versus pleomorphic adenoma of minor salivary glands: resolution of a diagnostic dilemma by immunohistochemical analysis with glial fibrillary acidic protein. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 2001;91:194–199.
Cunha IW, Lopes A, Falzoni R, et al. Sarcomas often express constitutive nitric oxide synthases (NOS) but infrequently inducible NOS. Appl Immunohistochem Mol Morphol 2006;14:404–410.
Rocha RM, Nunes CB, Rocha GFS, et al. Rabbit monoclonal antibodies show higher sensitivity than mouse monoclonals for estrogen and progesterone receptor evaluation in breast cancer by immunohistochemistry. Pathol Res Pract 2008;204:655–662.
Pfaffl MW . A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 2001;29:e45.
Vandesompele J, De Preter K, Pattyn F, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 2002, 3:RESEARCH0034.
Acknowledgements
FAPESP/CEPID: 98/14335.
Author information
Authors and Affiliations
Corresponding author
Additional information
Disclosure/conflict of interest
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Santos, G., Carvalho, K., Falzoni, R. et al. Glial fibrillary acidic protein in tumor types with cartilaginous differentiation. Mod Pathol 22, 1321–1327 (2009). https://doi.org/10.1038/modpathol.2009.99
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2009.99