Abstract
HER-2/neu gene amplification, found in certain subtypes of (breast-) cancers, is an independent prognostic factor of poor outcome and determines eligibility for systemic treatment with trastuzumab. TopoIIα (TOP2A) gene amplification seems to be predictive of response to a class of cytostatic agents called TopoII inhibitors, which include the anthracyclines. The observed increased efficacy of anthracyclines in HER2-positive tumors is thought to arise from the close proximity of both genes on chromosome 17, where the TopoII amplification status will determine the anthracycline sensitivity. This study aimed to validate a new polymerase chain reaction-based test, called multiplex ligation-dependent probe amplification (MLPA), as a simple and quick method to simultaneously assess HER-2/neu and TopoIIα gene amplification status in paraffin-embedded breast cancer samples. To this end, MLPA results were compared with TopoIIα, HER2 chromogenic in situ hybridization (CISH). We also assessed TopoIIα protein expression by immunohistochemistry. Of 353 patients, 9% showed TopoIIα amplification by MLPA and 13% of patients were HER2 amplified. TopoIIα amplification was seen in 42% of HER2-amplified cases and showed no high level amplification without HER2 amplification. Eleven patients displayed TopoIIα loss (3%). Concordance between MLPA and CISH was 91% for TopoIIα and 96% for HER2. Correlation between amplification and overexpression of TopoIIα was significant (P=0.035), but amplification did not always predict protein overexpression. Loss of the TopoIIα gene was almost never associated with loss of its protein. In conclusion, MLPA is an easy and accurate method to simultaneously detect breast cancer HER-2/neu and TopoIIα copy number status in paraffin-embedded tissue, and thus an attractive supplement or alternative to CISH.
Similar content being viewed by others
Main
The topoisomerase IIα (TopoIIα, TOP2A) gene is located at chromosome 17q21.2 and encodes a 170 kDa protein that has a key role in cell division by controlling and modifying the topological status of DNA.1 Furthermore, TopoIIα is the direct molecular target of TopoII inhibitors including anthracyclines, which are among the most powerful cytostatic agents in the treatment of invasive breast cancer. The binding of anthracyclines to TopoIIα is believed to stabilize the DNA double-strand breaks created by TopoIIα, leading to apoptosis. The TopoIIα gene is located next to the locus of the HER2 (human epidermal growth factor receptor 2) gene, a proto-oncogene belonging to the EGFR family. The HER2 gene encodes for a 185-kDa transmembrane glycoprotein, and overexpression of the protein is associated with poor prognostic as a consequence of increased cell proliferation, angiogenesis, invasive growth, and resistance to apoptosis. The HER2 gene is amplified and overexpressed in 10–30% of breast cancers, in which it has an important role in oncogenesis.2, 3 The HER2 protein is a direct target of trastuzumab (HerceptinR), a humanized monoclonal antibody that has been approved for the systemic treatment of both primary and metastatic breast cancer.4, 5, 6 With regard to the sensitivity of HER2-positive breast cancer, a number of studies have suggested an association with increased benefit of anthracycline-containing regimens. As a molecular basis for this association seems difficult to grasp, it has been suggested that the increased sensitivity of HER2/neu-positive breast cancer is a result from the proximity of the TopoIIα gene to the HER2 gene.7 Overall, TopoIIα amplification is considered to be an uncommon event in breast cancer, with a prevalence of approximately 5–10%.8, 9 Co-amplification of HER2 and TopoIIα is seen in approximately 40% of HER2-amplified breast cancer patients10, 11 and results of—mainly retrospectively obtained—data seem to underline the hypothesis that TopoIIα and not HER2 overexpression is the ultimate predictor of the response to anthracyclines.10, 12, 13, 14 Measurement of TopoIIα in the tumor could therefore potentially be useful in selecting the patients for treatment with TopoII inhibitors, including anthracyclines. Expression of TopoIIα protein has, however, not been shown to reliably predict response to anthracyclines, despite the fact that it is the direct target for these compounds.11, 15, 16, 17 In contrast, evaluation of TopoIIα gene copy number appears to be a good predictor of response to TopoIIα inhibitors.18, 19, 20 Furthermore, contrary to HER2, TopoIIα amplification has shown an inconsistent correlation with TopoIIα protein expression,21, 22 mainly because TopoIIα protein is highly dependent on the stage of the cell cycle and proliferation rate.
Recently, we introduced HER2 amplification detection in breast cancer by multiplex ligation-dependent probe amplification (MLPA). MLPA kits contain probes for up to 45 different targets allowing copy number assessment of different genes in the same PCR.23 MLPA requires only small quantities of short DNA fragments, which makes it very suitable for analysis of paraffin-embedded material. In previous studies using MLPA, we obtained promising results for HER2 in comparison with immunohistochemistry,24 fluorescence in situ hybridization and chromogenic in situ hybridization (CISH).25 As the applied HER2 kit also contains a TopoIIα probe, we set out to test MLPA as a new method to simultaneously assess HER2 and TopoIIα gene amplification status in a large group of breast cancer patients and to validate MLPA results with CISH in a subgroup of these patients. In addition, we investigated the correlation between TopoIIα protein expression levels and gene amplification status on tissue micro arrays, using immunohistochemistry and CISH, respectively.
Materials and methods
Patient Material
From a previously used study cohort (n=518), collected between November 2004 and June 2006 at the Department of Pathology of the University Medical Centre in Utrecht,25 353 consecutive tissue samples of invasive breast cancer patients were randomly selected. First, all tissue samples were analyzed by MLPA to determine HER2 and TopoIIα gene amplification status. For TopoIIα CISH and immunohistochemistry, tissue microarrays were constructed from the original paraffin-embedded tumor blocks (n=315) using published guidelines.26 In this study, the use of left over material was approved by the Tissue Science Committee of the UMC Utrecht.
Multiplex Ligation-Dependent Probe Amplification
Invasive tumor areas were harvested from 4 μm thick paraffin sections by dissection with a scalpel (using at least 1 cm2 tumor tissue) and DNA was isolated by 1 h incubation in proteinase K (10 mg/ml; Roche, Almere, The Netherlands) at 56°C followed by boiling for 10 min. This DNA solution (50–100 μl) was, after centrifugation, used in the MLPA analysis according the manufacturers’ instructions, using the P004-A1 HER2 kit (MRC Holland, Amsterdam, The Netherlands). This kit contains three probes for the HER2 gene, a probe for TopoIIα, 9 additional control probes for chromosome 17, and 25 control probes located on other chromosomes. Details of the probes in this kit can be found at http://www.mrc-holland.com. All tests were performed in duplicate on an ABI 9700 PCR machine (Applied Biosystems, Foster City, CA, USA). PCR products were analyzed on an ABI310 capillary sequencer. HER2 and TopoIIα gene copy numbers were normalized against the control probes in the kit, thereby excluding all chromosome 17 probes. The mean of all three HER2 probe peaks in duplicate (6 values) and the TopoIIα peak in duplicate (2 values) was calculated. If this mean value was below 0.7, TopoIIα or HER2 was considered lost, values between 0.7–1.5 were considered normal, values between 1.5 and 2.0 as low level amplified, and values >2.0 as HER2 or TopoIIα amplified. The 2.0 threshold was used in accordance with previous HER2 MLPA studies,24, 27 whereas the 1.5 threshold was empirically established during routine diagnostic application of MLPA kits for trisomy detection.
Chromogenic In Situ Hybridization
HER2 and TopoIIα CISH assays were performed on 4 μm thick paraffin serial tissue array sections using the SPoT-Light HER2 or TopoIIα kits (Zymed, San Francisco, CA, USA) according to the manufacturers’ instructions. First, sections were baked overnight at 56°C and deparaffinized in xylene and alcohol 100%. For HER2 and TopoIIα, the slides were then boiled in pretreatment buffer for 15 min, followed by enzymatic digestion at room temperature (RT) for 10 min (Zymed). Then, slides were dehydrated with graded alcohols. After 20 min of air-drying, the digoxigenin-labeled TopoIIα or HER2 probes were applied to the slides. Then, the sections were denatured on a hot plate (95°C) for 5 min and hybridization was carried out overnight at 37°C. After hybridization, appropriate stringency washes at 80°C were performed, followed by blocking with 3% hydrogen peroxide and CAS block (Zymed). Subsequently, the slides were incubated with mouse-anti-digoxigenin antibody (Zymed) for 30 min at RT and goat-anti-mouse antibody conjugated with horseradish peroxidase (HRP) for 30 min at RT. This was followed by diaminobenzidine (DAB) development for 30 min and counterstaining with hematoxylin. Finally, sections were dehydrated and mounted in Histomount (Zymed). A positive control was included in each CISH run and consisted of paraffin sections of a case known to be TopoIIα/HER2 amplified by CISH. At least 30 preferably non-overlapping nuclei in every tumor sample were scored by two blinded observers to determine the number of HER2 and TopoIIα signals. Amplification was defined to be present when large peroxidase-positive intra-nuclear clusters (or >10 individual small signals) were detected in at least 50% of tumor cells. The presence of small peroxidase-positive intra-nuclear clusters (or 6–10 individual small signals) was considered low level amplified. One to five individual small signals were scored as HER2/TopoIIα non-amplified.
Immunohistochemistry
Immunohistochemistry was performed using a mouse monoclonal antibody against the TopoIIα protein (clone Ki-S1, DAKO, Glostrup, Denmark) on 4 μm thick sections from neutral-buffered formaldehyde-fixed tissue array blocks. First, sections were baked overnight at 56°C, deparaffinized and rehydrated. The slides were then blocked in 3% hydrogen peroxide for 15 min and boiled in EDTA buffer (pH 9.0) for 20 min. After washing in 0.05% PBS Tween, the slides were incubated with the primary antibody at a dilution of 1/200 for 60 min at RT. Detection was performed with Envision (Dako, Glostrup, Denmark) using an HRP-conjugated secondary antibody followed by DAB development. The percentage of strongly positive nuclei was estimated (weakly positive nuclei were ignored). The median percentage of stained cells was 2%, we therefore defined >2% as overexpression. Immunohistochemistry expression was analyzed by one experienced (blinded) breast pathologist (PJvD) and at least 30 nuclei were scored.
Statistics
Results obtained with MLPA and CISH were compared by cross tables using SPSS for Windows and the concordance percentages were calculated. Correlations between continuous and categorical variables were performed with the non-parametric Mann–Whitney U-test. Correlations between categorical variables were performed using the χ2-test. P-values below 0.05 were considered significant.
Results
Multiplex Ligation-Dependent Probe Amplification
Table 1 shows the frequencies of TopoIIα and HER2 amplification. The TopoIIα gene was low level amplified in 7% of cases and highly amplified in 8 cases (2%), adding up to a total of 33/353 (9%) cases with amplification. HER2 was low level amplified in 10/353 cases (3%) and highly amplified in 34/353 cases (10%), adding up to a total of 44/353 (13%) of amplified cases.
Co-amplification with TopoIIα was seen in 42% of HER2-amplified cases (including both low and high levels). There was no high level amplification of TopoIIα without HER2 amplification. However, in some cases we found a low level amplification of TopoIIα without amplification of HER2. As to comparative copy numbers in co-amplified tumors, HER2 was often amplified at a higher level than TopoIIα within the same tumor.
Eleven cases (3%) were deleted for TopoIIα, all having a normal HER2 status.
Chromogenic In Situ Hybridization
TopoIIα and HER2 CISH were performed on 284 patients who were analyzed by MLPA (see Table 2). For TopoIIα we found concordance in 259 out of 284 (91%) of these patients. Most discordance was found in cases scored as low level by MLPA. Only 5/25 of these cases were confirmed to be TopoIIα amplified by CISH, and the other 20 cases were scored normal by CISH. All MLPA highly amplified cases were confirmed by CISH, although two of these cases only showed a low level amplification by CISH. Of the non-amplified cases by MLPA, 249 (99%) were concordant with CISH, whereas two non-amplified cases were scored low level amplified by CISH.
For HER2, 273/284 (96%) cases were concordant between MLPA and CISH. Concordance was highest in MLPA amplified (27/28) and non-amplified (244/248) cases, whereas 4/7 MLPA low level cases were scored normal by CISH.
Sensitivity, specificity, positive-predictive value and negative-predictive value of MLPA for HER2 and TopoIIα were calculated and depicted in Table 4 using CISH results as gold standard, and by taking low level and high level amplifications together. When the cut-off was set at 1.8, the number of low level amplified patients was reduced significantly, thereby increasing the concordance between MLPA and CISH (as gold standard) for TopoIIα. Nevertheless, increasing the cut-off value lead to a decrease in sensitivity of MLPA for both genes.
Immunohistochemistry
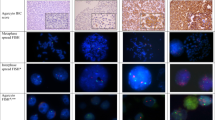
From tissue arrays containing cores of 315 patients, information for both immunohistochemistry and CISH was obtained for 265 patients. A strong positive nuclear staining for Topoisomerase IIα in 265 invasive breast tumors ranged from 0 to 90% of tumor cells. One hundred and seventeen cases (44%) showed overexpression. Topoisomerase IIα overexpression was significantly associated with TopoIIα amplification by MLPA (P=0.035), although 4/14 (29%) of amplified tumors did not overexpress the TopoIIα protein (Table 3, Figure 1). One patient showing amplification of TopoIIα by CISH was not analyzed by immunohistochemistry because there was not enough tissue left. Of the cases without TopoIIα amplification, 42% showed overexpression, in comparison with 71% for TopoIIα-amplified cases. There was also evidence of a difference (P=0.01) in the mean TopoIIα protein expression level for tumor samples with TopoIIα amplification by CISH (n=14, mean 28% immunohistochemistry positive) vs no TopoIIα gene amplification (n=251, mean 7% immunohistochemistry positive).
Correlation between gene amplification and protein expression in breast cancer as determined by chromogenic in situ hybridization and immunohistochemistry. Top left: Almost no TopoIIα protein expression is present. Top right: Large chromogenic in situ hybridization clusters indicate TopoIIα gene amplification in the same patient. Bottom left: strong TopoIIα protein expression is present in 5% of tumor cells. Bottom right: chromogenic in situ hybridization shows less than 5 signals per cell indicating no TopoIIα gene amplification.
Loss of the TopoIIα gene (n=15) was rarely (2/15) accompanied by absence of its protein, but rather by overexpression (7/15) although not significantly (P=0.421).
Discussion
The aim of this study was to test MLPA as a new method to simultaneously assess HER2 and TopoIIα gene amplification status in a large group of breast cancer patients, and to compare MLPA results with CISH data as gold standard in a selected group of patients. Of 353 patients analyzed by MLPA, 2% showed a high level amplification of the TopoIIα gene and 10% of patients manifested a high level amplification of the HER2 gene. When including low amplification, the percentages of amplification rose to 9 and 13%, respectively. For HER2 this is lower than the 20–30% positivity that has generally been described in the literature,2, 3, 28, 29 although several other studies have reported lower (10–18%) percentages30, 31, 32, 33, 34 as well. It is likely that many of the series in which higher HER2 overexpression/amplification frequencies were described have not been unselected, whereas frequencies below 20% have been reported before in unselected series. As our study group concerned consecutive patients, selection bias can be excluded. Furthermore, methodological variation is an unlikely explanation as the fraction of HER-2/neu-amplified cases by immunohistochemistry (10%, Moelans et al25) was similar. This implies that there may be geographic variations in HER-2/neu- and TopoIIα-amplification status. TopoIIα amplification has been described to be present in approximately 5–10% of the total population (about one-third of HER2-amplified tumors),11 which is consistent with our data (9%).
Co-amplification of HER2 and TopoIIα was seen in 42% of cases (low and high level) in line with previous studies that reported co-amplification rates of 32–57%.10, 11 We found no high level amplification of TopoIIα without HER2 amplification, in contrast with some studies that did find TopoIIα amplification with normal HER2 status.8, 35 However, in some cases we found a low level of TopoIIα gene amplification without any amplification of HER2, but this amplification could not be identified by CISH.
Copy numbers of HER2 were higher than those of TopoIIα, which in addition to the different frequency of amplification of these loci supports the concept that the HER2 gene is the hot spot for amplification on chromosome 17, with lower frequencies of amplification and lower level of amplification of the surrounding genes such as TopoIIα36 and other chromosome 17q genes included in the kit (as depicted in Figure 2). Nevertheless, the mechanism of amplification of the HER2 gene and surrounding loci is yet unknown. To which extent these co-amplified genes have an impact on response to the HER2-targeted treatment with trastuzumab is unknown.
Schematic representation of the HER-2/neu amplicon. The HER2 (ERBB2) core region as defined by Kauraniemi et al46 is indicated by dashed lines. Genes corresponding to the chromosome 17 probes included in the MLPA P004 kit are depicted above the chromosome and their (low plus high level) amplification frequencies (in %) as found in this study are depicted in the chart below.
Eleven patients showed a deletion for TopoIIα by MLPA (3%), which is consistent with literature, where overall prevalence of TopoIIα deletions in breast cancer has varied from 2 to 11% in different studies.10, 11, 37 In our study, none of these deletions was accompanied by an amplification of HER2. The significance of these deletions is still controversial, but contrary to what was previously thought,18 one study claimed that it may also predict benefit from treatment with TopoIIα inhibitors.8
In our study, tumors with gene amplification of Topoisomerase IIα showed evidence of greater expression of topoisomerase IIα protein than did other tumors (P=0.035), but 4/14 (29%) amplified tumors did not overexpress the TopoIIα protein. All four cases displayed low level amplification by CISH, and two of these four cases were also amplified by MLPA. Previous studies have revealed that, contrary to HER2, where gene amplification is almost always correlated with protein overexpression, TopoIIα gene amplification apparently does not always lead to protein overexpression.12, 21, 22 Other factors, specifically the tumor proliferation status, may interfere with the TopoIIα protein status as topoisomerase IIα is a marker of proliferation and topoisomerase IIα expression depends on the cell cycle status.
We found, similar to a large previous study,25 a high concordance between amplification status by MLPA and CISH, which indicates that MLPA is a reliable test for detection of HER-2/neu and TopoIIα amplification. One can even wonder whether MLPA would be suitable as a pre-screening tool alternative to the Hercep test (HER2 immunohistochemistry). Indeed, MLPA is not only easy but also cheaper than CISH. Consumables costs are €11 per reaction compared with €70 per reaction for HER2 CISH and €56 per reaction for TopoIIα CISH. Furthermore, MLPA is more quantitative than immunohistochemistry allowing more straightforward interpretation, and in the same analysis several genes that are important in therapy selection and/or prognosis, such as TopoIIα, can be tested for amplification. Given the inherent molecular complexity of the malignant process, it seems unlikely that the assay of a single marker, regardless of methodology, will ever give us the complete answer as to the response to targeted therapeutics.
Concordance with CISH for TopoIIα was 91%, for HER2 96%. This difference could be due to a more accurate estimation of HER2 status based on three probes instead of only one for TopoIIα in the current kit, indicating that the kit would benefit from more TopoIIα probes. More MLPA probes for TopoIIα could make a more accurate estimate of whether a sample is low level or not amplified. Concordance between MLPA and CISH for HER2 and TopoIIα was highest in MLPA amplified (96 and 100%, respectively) and non-amplified cases (98 and 99%, respectively). For MLPA low-level amplified cases, concordance was low (50 and 16%). However, low-level HER2 amplification only occurs in 1–3% of the general population and in 4–25% of the critical group of immunohistochemistry 2+ carcinomas.38 These low-level amplified cases probably do not respond as well to HER2-directed therapy as patients showing high level amplifications.39 Preliminary data from the NSABP B-31 trial, however, suggest that there is a limited subset of patients with tumors that are fluorescence in situ hybridization negative and graded less than immunohistochemistry 3+ that do achieve significant benefit (P=0.03) from adjuvant trastuzumab.40 We therefore re-analyzed our MLPA results with higher cut-off values (than 1.5) between non-amplified and low-level amplified cases (see Table 4), which increased the positive predictive value and specificity but decreased the sensitivity of MLPA for both genes. Next to the number of probes and the choice of the cut-off value, another explanation of discrepancies could be the non-morphological aspect of MLPA. Small-amplified clones may be obscured by background non-amplified cells and thereby missed by MLPA. Careful manual microdissection is able to resolve some of the discrepancies, but is not necessary in routine practice and only advisable when tumor percentage is very low (<30%) or extensive ductal carcinoma in situ is present.41 On the other hand, there were also cases with amplification by MLPA, while CISH was normal. This may be partly due to a lack of sensitivity by CISH for low level amplification.
These data show that MLPA is suited to detect amplification (as well as deletion) of HER2 and TopoIIα in breast cancer patients in one test. Both HER2 and TopoIIα gene alternations have independently been associated with an increased responsiveness to anthracycline-containing chemotherapy regimens relative to non-anthracyline regimens,42, 43 indicating that measurements of alternations of both genes can guide in the selection of anthracyline-containing regimens. Furthermore, this MLPA kit contains probes to several other chromosome 17 loci (see Figure 2) and can thereby easily determine chromosome 17 polysomy, likely better than using a single in situ hybridization centromere probe, and easier than additional in situ hybridization probes targeted to other chromosome 17 loci.44 This is even more an advantage, as recently the definition of chromosome 17 polysomy based on CEP17 only is found most questionable.45
In conclusion, MLPA is an easy and cheaper method to simultaneously detect breast cancer polysomy 17, HER-2/neu and TopoIIα amplification in small quantities of short fragmented DNA extracted from paraffin blocks, and is thereby a good supplementary or even alternative technique to in situ hybridization.
References
Berger JM, Gamblin SJ, Harrison SC, et al. Structure and mechanism of DNA topoisomerase II. Nature 1996;379:225–232.
Slamon DJ, Clark GM, Wong SG, et al. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science 1987;235:177–182.
Slamon DJ, Godolphin W, Jones LA, et al. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science 1989;244:707–712.
Slamon DJ, Leyland-Jones B, Shak S, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med 2001;344:783–792.
Piccart-Gebhart MJ, Procter M, Leyland-Jones B, et al. Trastuzumab after adjuvant chemotherapy in HER2-positive breast cancer. N Engl J Med 2005;353:1659–1672.
Romond EH, Perez EA, Bryant J, et al. Trastuzumab plus adjuvant chemotherapy for operable HER2-positive breast cancer. N Engl J Med 2005;353:1673–1684.
Tanner M, Isola J, Wiklund T, et al. Topoisomerase IIalpha gene amplification predicts favorable treatment response to tailored and dose-escalated anthracycline-based adjuvant chemotherapy in HER-2/neu-amplified breast cancer: Scandinavian Breast Group Trial 9401. J Clin Oncol 2006;24:2428–2436.
Knoop AS, Knudsen H, Balslev E, et al. Retrospective analysis of topoisomerase IIa amplifications and deletions as predictive markers in primary breast cancer patients randomly assigned to cyclophosphamide, methotrexate, and fluorouracil or cyclophosphamide, epirubicin, and fluorouracil: Danish Breast Cancer Cooperative Group. J Clin Oncol 2005;23:7483–7490.
Di Leo A, Gancberg D, Larsimont D, et al. HER-2 amplification and topoisomerase IIalpha gene aberrations as predictive markers in node-positive breast cancer patients randomly treated either with an anthracycline-based therapy or with cyclophosphamide, methotrexate, and 5-fluorouracil. Clin Cancer Res 2002;8:1107–1116.
Slamon D, Eiermann W, Robert N . Phase III randomized trial comparing doxorubicin and cyclophosphamide followed by docetaxel (ACT) with doxorubicin and cyclophosphamide followed by docetaxel and trastuzumab (ACTH) with docetaxel, carboplatin and trastuzumab (TCH) in Her2-positive early breast cancer patients: BCIRG006 study. [abstract]. Breast Cancer Res Treat 2005;94:S5, 2005 (Suppl 1; abstract 1).
Mano MS, Rosa DD, de AE, et al. The 17q12-q21 amplicon: Her2 and topoisomerase-IIalpha and their importance to the biology of solid tumours. Cancer Treat Rev 2007;33:64–77.
Coon JS, Marcus E, Gupta-Burt S, et al. Amplification and overexpression of topoisomerase IIalpha predict response to anthracycline-based therapy in locally advanced breast cancer. Clin Cancer Res 2002;8:1061–1067.
Pritchard KI, Messersmith H, Elavathil L, et al. HER-2 and topoisomerase II as predictors of response to chemotherapy. J Clin Oncol 2008;26:736–744.
Di Leo A, Biganzoli L, Claudino W, et al. Topoisomerase II alpha as a marker predicting anthracyclines’ activity in early breast cancer patients: ready for the primetime? Eur J Cancer 2008;44:2791–2798.
Petit T, Wilt M, Velten M, et al. Comparative value of tumour grade, hormonal receptors, Ki-67, HER-2 and topoisomerase II alpha status as predictive markers in breast cancer patients treated with neoadjuvant anthracycline-based chemotherapy. Eur J Cancer 2004;40:205–211.
Di Leo A, Larsimont D, Gancberg D, et al. HER-2 and topo-isomerase IIalpha as predictive markers in a population of node-positive breast cancer patients randomly treated with adjuvant CMF or epirubicin plus cyclophosphamide. Ann Oncol 2001;12:1081–1089.
Jarvinen TA, Holli K, Kuukasjarvi T, et al. Predictive value of topoisomerase IIalpha and other prognostic factors for epirubicin chemotherapy in advanced breast cancer. Br J Cancer 1998;77:2267–2273.
Jarvinen TA, Tanner M, Rantanen V, et al. Amplification and deletion of topoisomerase IIalpha associate with ErbB-2 amplification and affect sensitivity to topoisomerase II inhibitor doxorubicin in breast cancer. Am J Pathol 2000;156:839–847.
Asano T, Kleinerman ES, Zwelling LA, et al. Adenovirus-mediated human topoisomerase IIalpha gene transfer increases the sensitivity of etoposide-resistant human and mouse breast cancer cells. Acta Oncol 2005;44:240–247.
Nielsen KV, Ejlertsen B, Moller S, et al. The value of TOP2A gene copy number variation as a biomarker in breast cancer: Update of DBCG trial 89D. Acta Oncol 2008;47:725–734.
Durbecq V, Paesmans M, Cardoso F, et al. Topoisomerase-II alpha expression as a predictive marker in a population of advanced breast cancer patients randomly treated either with single-agent doxorubicin or single-agent docetaxel. Mol Cancer Ther 2004;3:1207–1214.
Mueller RE, Parkes RK, Andrulis I, et al. Amplification of the TOP2A gene does not predict high levels of topoisomerase II alpha protein in human breast tumor samples. Genes Chromosomes Cancer 2004;39:288–297.
Schouten JP, McElgunn CJ, Waaijer R, et al. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res 2002;30:e57.
Purnomosari D, Aryandono T, Setiaji K, et al. Comparison of multiplex ligation dependent probe amplification to immunohistochemistry for assessing HER-2/neu amplification in invasive breast cancer. Biotech Histochem 2006;81:79–85.
Moelans CB, de Weger RA, van Blokland MT, et al. HER-2/neu amplification testing in breast cancer by multiplex ligation-dependent probe amplification in comparison with immunohistochemistry and in situ hybridization. Cell Oncol 2009;31:1–10.
Packeisen J, Korsching E, Herbst H, et al. Demystified.tissue microarray technology. Mol Pathol 2003;56:198–204.
Moerland E, van Hezik RL, van der Aa TC, et al. Detection of HER2 amplification in breast carcinomas: comparison of Multiplex Ligation-dependent Probe Amplification (MLPA) and Fluorescence In situ Hybridization (FISH) combined with automated spot counting. Cell Oncol 2006;28:151–159.
Owens MA, Horten BC, Da Silva MM . HER2 amplification ratios by fluorescence in situ hybridization and correlation with immunohistochemistry in a cohort of 6556 breast cancer tissues. Clin Breast Cancer 2004;5:63–69.
Garcia-Caballero T, Menendez MD, Vazquez-Boquete A, et al. HER-2 status determination in breast carcinomas. A practical approach. Histol Histopathol 2006;21:227–236.
Dandachi N, Dietze O, Hauser-Kronberger C . Chromogenic in situ hybridization: a novel approach to a practical and sensitive method for the detection of HER2 oncogene in archival human breast carcinoma. Lab Invest 2002;82:1007–1014.
Perez EA, Roche PC, Jenkins RB, et al. HER2 testing in patients with breast cancer: poor correlation between weak positivity by immunohistochemistry and gene amplification by fluorescence in situ hybridization. Mayo Clin Proc 2002;77:148–154.
Ellis CM, Dyson MJ, Stephenson TJ, et al. HER2 amplification status in breast cancer: a comparison between immunohistochemical staining and fluorescence in situ hybridisation using manual and automated quantitative image analysis scoring techniques. J Clin Pathol 2005;58:710–714.
Dolan M, Snover D . Comparison of immunohistochemical and fluorescence in situ hybridization assessment of HER-2 status in routine practice. Am J Clin Pathol 2005;123:766–770.
Marla S, Roxburgh P, Burton P, et al. HER2 positive early breast cancers: tumour demographics and trastuzumab therapy in the real-world [abstract]. Cancer Res 2009;69 (Suppl 2):11.
Olsen KE, Knudsen H, Rasmussen BB, et al. Amplification of HER2 and TOP2A and deletion of TOP2A genes in breast cancer investigated by new FISH probes. Acta Oncol 2004;43:35–42.
Jarvinen TA, Tanner M, Barlund M, et al. Characterization of topoisomerase II alpha gene amplification and deletion in breast cancer. Genes Chromosomes Cancer 1999;26:142–150.
Bhargava R, Lal P, Chen B . HER-2/neu and topoisomerase IIa gene amplification and protein expression in invasive breast carcinomas: chromogenic in situ hybridization and immunohistochemical analyses. Am J Clin Pathol 2005;123:889–895.
van de Vijver M, Bilous M, Hanna W . Chromogenic in-situ hybridisation (CISH) compared with FISH and IHC for detection of HER2 gene amplification: an international validation ring study [abstract]. Breast Cancer Res Treat 2003;82:S75 (suppl 1, abstract 324).
Lewis F, Jackson P, Lane S, et al. Testing for HER2 in breast cancer. Histopathology 2004;45:207–217.
Paik S, Kim C, Jeong J . Benefit from adjuvant trastuzumab may not be confined to patients with IHC 3+ and/or FISH-positive tumors: central testing results from NSABP B-31 [abstract]. Proc Am Soc Clin Oncol 2007;25 (Suppl 18):511.
Moelans CB, de Weger RA, Ezendam C, et al. HER-2/neu amplification testing in breast cancer by Multiplex Ligation-dependent Probe Amplification: influence of manual- and laser microdissection. BMC Cancer 2009;9:4.
O’Malley FP, Chia S, Tu D, et al. Topoisomerase II alpha and responsiveness of breast cancer to adjuvant chemotherapy. J Natl Cancer Inst 2009;101:644–650.
Slamon DJ, Press MF . Alterations in the TOP2A and HER2 genes: association with adjuvant anthracycline sensitivity in human breast cancers. J Natl Cancer Inst 2009;101:615–618.
Troxell ML, Bangs CD, Lawce HJ, et al. Evaluation of Her-2/neu status in carcinomas with amplified chromosome 17 centromere locus. Am J Clin Pathol 2006;126:709–716.
Yeh I-T, Martin M, Robetorye R, et al. ‘Polysomy 17’ by FISH for HER2 in breast cancer is a misnomer: results of chromosome 17 analysis by array comparative genomic hybridization [abstract]. Cancer Res 2009;69 (Suppl 2):11.
Kauraniemi P, Kuukasjarvi T, Sauter G, et al. Amplification of a 280-kilobase core region at the ERBB2 locus leads to activation of two hypothetical proteins in breast cancer. Am J Pathol 2003;163:1979–1984.
Author information
Authors and Affiliations
Corresponding author
Additional information
Disclosure/conflict of interest
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Moelans, C., de Weger, R., van Blokland, M. et al. Simultaneous detection of TOP2A and HER2 gene amplification by multiplex ligation-dependent probe amplification in breast cancer. Mod Pathol 23, 62–70 (2010). https://doi.org/10.1038/modpathol.2009.136
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2009.136