Abstract
Macrophages (mφ) are essential for intestinal homeostasis and the pathology of inflammatory bowel disease (IBD), but it is unclear whether discrete mφ populations carry out these distinct functions or if resident mφ change during inflammation. We show here that most resident mφ in resting mouse colon express very high levels of CX3CR1, are avidly phagocytic and MHCIIhi, but are resistant to Toll-like receptor (TLR) stimulation, produce interleukin 10 constitutively, and express CD163 and CD206. A smaller population of CX3CR1int cells is present in resting colon and it expands during experimental colitis. Ly6ChiCCR2+ monocytes can give rise to all mφ subsets in both healthy and inflamed colon and we show that the CX3CR1int pool represents a continuum in which newly arrived, recently divided monocytes develop into resident CX3CR1hi mφ. This process is arrested during experimental colitis, resulting in the accumulation of TLR-responsive pro-inflammatory mφ. Phenotypic analysis of human intestinal mφ indicates that analogous processes occur in the normal and Crohn's disease ileum. These studies show for the first time that resident and inflammatory mφ in the intestine represent alternative differentiation outcomes of the same precursor and targeting these events could offer routes for therapeutic intervention in IBD.
Similar content being viewed by others
Introduction
Selective inertia of intestinal macrophages (mφ) is one of the principal mechanisms preventing the inappropriate reactions to commensal microbes that cause inflammatory bowel diseases (IBD) such as Crohn's disease.1 Despite being actively phagocytic and bactericidal, resident mucosal mφ fail to produce pro-inflammatory mediators in response to stimuli such as Toll-like receptor (TLR) ligands.2 Instead they have a number of crucial homeostatic functions, acting as non-inflammatory scavengers of bacteria, as well as assisting the maintenance of regulatory T cells and promoting epithelial cell renewal, via the production of interleukin 10 (IL10) and PGE2, respectively.2, 3, 4 However, mφ are also important effectors of pathology in IBD, producing inflammatory mediators such as tumor necrosis factor α (TNFα), IL1, IL6, and nitric oxide.5 Elucidating how mucosal mφ can be capable of such different functions would be important both for developing new therapeutics and understanding intestinal physiology.
It has been assumed that distinct populations of intestinal mφ are responsible for their functional plasticity under different conditions,6 an idea consistent with the paradigm that there are “resident” and “inflammatory” lineages of monocytes.2 However, this possibility has not been studied in any depth in the intestine. Furthermore, previous experiments are often difficult to interpret, because they pre-gated for “antigen presenting cells” and used phenotypic markers such as CD11b, CD11c, MHCII, and F4/80 that are now recognized not to discriminate between different populations of mononuclear phagocytes in tissues like the gut.7
Recent studies have refined the characterization of intestinal mononuclear phagocytes (MP), based on the mutually exclusive expression of the αE integrin (CD103) and the chemokine receptor CX3CR1.8, 9, 10 On the basis of this work, it is believed that CD103− expressing CD11c+MHCII+ LP cells are bona fide dendritic cells (DC) that migrate from the LP to the mesenteric lymph node and have a unique capacity to prime naïve T cells. However, the phenotype and function of CX3CR1+ MP is less clear. Although sometimes considered to be a CD103− subset of DC, they do not migrate to mesenteric lymph node, cannot prime naïve T cells, and are now thought to be tissue-resident mφ.9, 10 Recently, the level of CX3CR1 expression has been suggested to define heterogeneity within the CX3CR1+ population in mouse colon. Whereas the CX3CR1hi subset appears to be a group of tissue-resident mφ, those expressing intermediate amounts (CX3CR1int) were inherently inflammatory and predominated during experimental colitis.11 However, it was not clear from this work if these subsets represented distinct lineages of mφ, and their origins and relationship to other MP remain to be determined.
Here, we have applied multiparameter flow cytometry and lineage-tracking strategies to characterize CX3CR1-expressing MP cells in healthy and inflamed mouse and human colon. This revealed unsuspected heterogeneity of these cells and we present evidence that newly arrived inflammatory monocytes differentiate in situ into IL10-producing anti-inflammatory CX3CR1hi-resident mφ through a number of transitional stages. During intestinal inflammation, this differentiation process is disrupted, leading to the accumulation of TLR-responsive CX3CR1int mφ that produce inflammatory mediators. Thus resident and inflammatory mφ in the intestine represent alternative differentiation outcomes of the same precursor.
Results
Phenotypic characterization of CX3CR1+ mononuclear phagocyte populations in the colon
To characterize mφ in steady-state mouse colon, we developed strategies to analyze multiple markers on total colonic mucosal cells without prior selection by density gradients or markers of putative APC.
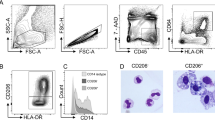
Three distinct populations of CD11b+ cells were identifiable among live CD45+ cells in CX3CR1+/gfp mice: CX3CR1−, CX3CR1int, and CX3CR1hi (Figure 1a; Supplementary Figure S1 online). The CX3CR1− fraction of cells was heterogeneous, containing SiglecFhi F4/80lo SSChi eosinophils, CD11chi MHCIIhi F4/80− DC, of which ∼20% expressed CD103, plus some Ly6Ghi neutrophils (Figure 1b; Supplementary Figure S1 online and data not shown).
Phenotypic characterization of colonic mononuclear phagocytes. (a) Doublets were excluded from colonic digests from CX3CR1gfp/+ mice on the basis of FSC-A and FSC-H, and live leukocytes were selected as being 7-AAD− CD45+. The resulting cells were then analyzed for CD11b and CX3CR1-green fluorescent protein (GFP) expression. (b) Histograms show the expression of the indicated cell surface markers on the CX3CR1−, CX3CR1int, and CX3CR1hi subsets. (c) Expression of Ly6C, MHCII, F4/80, and CD11c on CX3CR1int (top panels) and CX3CR1hi CD11b+ cells (bottom panels). (d) Morphological assessment of fluorescence-activated cell-sorted P1, P3, and P4 cells (× 200). (e) Phagocytic activity of colonic MP cell subsets, as measured by the uptake of pHrodo E. coli bioparticles for 15 min at 37 °C (black lines) or at 4 °C as a control (shaded histograms). Representative pHrodo dye fluorescence of CX3CR1-based P1 to P5 and CD103+ DC. Mean ΔMFI (mean fluorescence intensity (MFI) of pHrodo dye fluorescence at 37 °C-MFI at 4 °C control) for populations 1 to 4 + 1 s.d. for 4–5 mice is shown. Representation of three individual experiments (***P<0.001).
The majority CX3CR1hi cells uniformly expressed high levels of F4/80 and MHCII, but lacked CD103, SiglecF, or Ly6C. Most expressed CD11c, but at variable levels (Figure 1b and c) and all were CD64hi (Supplementary Figure S1d online), a marker that discriminates mφ and monocyte-derived DC from conventional DC.12 Significantly, they expressed higher levels of green fluorescent protein (GFP) than any other myeloid cells examined (Supplementary Figure S1 online). These CX3CR1hi cells had typical features of resident mφ, being large with abundant foamy cytoplasm and prominent cytoplasmic vacuoles, as well as high phagocytic activity (Figure 1d and e).
The smaller population of CX3CR1int cells was heterogeneous, with four subsets being identifiable based on MHCII, CD11c, Ly6C, and F4/80 expression (Figure 1b and c; Supplementary Figure S1g online). Population 1 (P1) expressed high levels of Ly6C, but low levels of F4/80 and it lacked MHCII, CD11c, or CD103. Together with their morphological appearance and lack of phagocytic activity (Figure 1d and e), P1 cells appear identical to Ly6Chi “inflammatory” monocytes in blood. P2 cells were similar to P1, except that they expressed MHCII. The remaining CX3CR1int cells were more numerous, expressed high levels of MHCII, and lacked Ly6C. These could be further subdivided into F4/80+CD11c−/lo (P3) and F4/80−CD11c+ subsets (P5) (Figure 1c; Supplementary Figure S1g online). The larger, F4/80+CD11c−/lo fraction was morphologically similar to the CX3CR1hi population (designated P4) (Figure 1d and e), consistent with them being relatively mature tissue mφ. All the cells in P1–P4 expressed CD64, but this was upregulated on P2 and P3 compared with P1, and increased further on P4 (Supplementary Figure S1d online). The F4/80negCD11c+ cells in P5 were CD64− and expressed lower levels of CX3CR1 than P3 (Supplementary Figure S1d and e online). Like CD103+ DC, they were non-phagocytic (Figure 1e) and their numbers expanded dramatically after in vivo administration of flt3L suggesting these are bona fide DC (Supplementary Figure S2 online). The F4/80+CD11c− cells in P3 did not expand in response to flt3L, nor did the P1, P2, or CX3CR1hi P4 cells, supporting the idea that these are part of the mφ lineage.
These data show that true intestinal mφ can only be identified accurately using a combination of markers that exclude DC and other leukocytes. They also reveal a novel phenotypic continuum among these cells ranging from monocytes at one end to mature mφ at the other.
Gene expression by mφ subsets in resting intestine
Quantitative PCR analysis of sorted subsets showed that progressive alterations in gene expression accompanied the phenotypic changes we had observed. Confirming the fluorescence-activated cell sorting (FACS) analysis, CX3CR1 messenger RNA (mRNA) levels increased from the P1 stage until they were maximal in CX3CR1hi mφ (P4) (Figure 2a). In parallel, there were incremental increases in the expression of CD163, CD206, and TGFβR2, markers associated with anti-inflammatory mφ.13 Conversely, the pro-inflammatory genes CCR2, IL6, and iNOS were highly expressed in P1, but were then extinguished by P4 (Figure 2a). There was no upregulation of arginase or vascular endothelial growth factor (Figure 2a, data not shown).
Transcriptional and kinetic analysis of colonic CX3CR1-defined myeloid cells. (a) Quantitative reverse transcription PCR (qRT-PCR) quantitation of CX3CR1, CD163, CD206, TGFβR2, CCR2, interleukin 6 (IL6), and tumor necrosis factor α (TNFα) messenger RNA by fluorescence-activated cell sorted P1, P2, P3, and P4 from colonic mucosa. Results shown are mean expression relative to cyclophilin A using the 2−ΔC(t) method. Means of three independent experiments using pooled cells from 12 mice per experiment. (b, c) CX3CR1+/gfp mice received 1 mg 5-bromo-2-deoxyuridine (BrdU) intraperitoneally and 3, 12, 24, and 48 h later, BrdU uptake was assessed in Ly6Chi blood and bone marrow monocytes (CD11b+Ly6ChiCX3CR1int) (b) and colonic CX3CR1int (P1–P3) and CX3CR1hi (P4) populations (c). Results are means of four mice per group.
These results support the idea that there is in situ differentiation of Ly6Chi monocytes into CX3CR1hi mφ in the resting colon, with stepwise acquisition of anti-inflammatory genes and downregulation of typical pro-inflammatory genes.
Ly6Chi monocytes give rise to resident Mφ subsets in resting colon
To explore further the idea that the different subsets of intestinal mφ might derive from local development of recently arrived monocytes, we performed 5-bromo-2-deoxyuridine (BrdU) pulse-chase studies to track Ly6Chi monocytes after release from the bone marrow (BM). At 3 h after a single pulse of BrdU, a significant proportion of Ly6Chi monocytes in BM had incorporated BrdU, but no BrdU+ cells could be found in the bloodstream, or among any of the colonic MP cell subsets (Figure 2b and c; Supplementary Figure S3a online). In contrast, 12 h after BrdU injection, BrdU+ Ly6Chi monocytes were now abundant in blood and BrdU+ cells were also seen among colonic P1 cells. All other subsets remained BrdU− at this time. By 24 h, BrdU+ cells were now apparent within the P2 population and these increased further at 48 h, by which time labeled cells were detectable in P3 and those in P1 were decreasing (Figure 2b and c; Supplementary Figure S3a online).
Thus the populations of CX3CR1int cells we identified in resting colon appear to be short-lived intermediaries in the local differentiation of Ly6Chi monocytes.
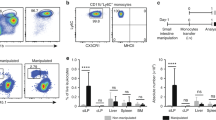
To test this hypothesis directly, we next examined the fate of adoptively transferred Ly6Chi monocytes in the steady-state colon using CCR2−/− mice as recipients, which have a selective defect in circulating Ly6Chi monocytes (Figure 3a). As these recipients do not express the CX3CR1-GFP reporter gene and we were unable to detect CX3CR1 reliably using antibodies, we developed an alternative gating strategy to characterize MP cells in non-CX3CR1-GFP strains (Supplementary Figure S4 online). This showed that resting CCR2−/− mice lack Ly6Chi monocytes (P1) and Ly6C+MHCII+ transitional cells (P2) in the colon, but have relative preservation of F4/80+MHCII+Ly6C− mφ (P3 and P4 in CX3CR1+/gfp mice) (Figure 3b).
Recruitment of Ly6Chi monocytes to colon of CCR2 null mice. (a) Representative staining and mean percentage of CD115+ Ly6Chi monocytes within the CD11b+ fraction of blood leukocytes in wild-type (WT) or CCR2−/− mice. (b) Representative Ly6C and MHCII expression on live-gated CD45+ CD11b+SSClow colon leukocytes and the absolute numbers of P1, P2, and P3/P4 subsets from resting WT or CCR2−/− mice. Results representative of at least three individual experiments with 3–4 mice in each group. (**P<0.01 and ***P<0.001). (c) Donor-derived cells (CD45.1+/CD45.2+ CX3CR1+/gfp) within the live-gated CD45+ CD11b+ fraction of colonic LP cells of CCR2−/− mice, 24 h, 48 h, 96 h, and 7 days after transfer of 2 × 106 fluorescence-activated cell-sorted Ly6Chi BM monocytes. (d) Expression of Ly6C, MHCII, and F4/80 on donor-derived cells isolated from recipient CCR2−/− colon at 24 h, 48 h, 96 h, and 7 days after transfer of Ly6Chi monocytes, compared with live-gated CD45+ CD11b+SSClo cells from wild-type (WT) colon (left panel). (e) Expression of CX3CR1-green fluorescent protein (GFP) by donor-derived cells in colon at 24 h, 48 h, 96 h, and 7 days after transfer. Gates for identifying CX3CR1int and CX3CR1hi cells were set using CD11b+ cells from resting CX3CR1+/gfp colon. (f) Donor-derived cells (CD45.1+/CD45.2+ CX3CR1+/gfp) among live-gated CD45+ CD11b+ cells in bloodstream of CCR2−/− mice, 24 h, 48 h, 96 h, and 7 days after transfer of Ly6Chi monocytes and in phosphate-buffered saline (PBS)-treated controls. Results are representative of two individual experiments with two recipient mice at each time point.
Purified CX3CR1+/gfp Ly6Chi BM monocytes (Supplementary Figure S5a online) were found in the colon of unmanipulated CCR2−/− recipients within 24 h after transfer, when all were CX3CR1int and ∼50% had acquired MHCII (Figure 3c and d). By 48 h, almost 90% of donor cells had upregulated MHCII, lost Ly6C, and showed some increase in CX3CR1 expression (Figure 3c–e). By 96 h after transfer virtually all donor monocytes had acquired MHCII, upregulated F4/80 and CD11c, and expressed high levels of CX3CR1 (Figure 3c–e). Donor-derived CX3CR1hi mφ could still be identified in the colon of recipient CCR2 KO mice 7 days after transfer, indicating at least some of these cells are relatively long lived. Importantly, donor Ly6Chi monocytes did not become MHCII+ CX3CR1hi in the bloodstream, or in other tissues of CCR2−/− recipients, including the lung (Figure 3f, data not shown). Consistent with previous studies,14, 15 adoptively transferred Ly6Clo monocytes could not be found in the colon of CCR2−/− or WT mice at any time, despite being obvious in the blood (Supplementary Figure S5b and c online).
Identical monocyte differentiation was seen using untreated CX3CR1gfp/+ mice and diphtheria toxin-treated CD11c-DTR mice as recipients (Supplementary Figures S5c and S6 online). These experiments show directly that Ly6Chi monocytes constantly enter the steady-state colon and mature into CX3CR1hi mφ through a series of CX3CR1int intermediaries.
Colonic inflammation modifies the composition of the local mφ pool
As Ly6Chi monocytes are conventionally associated with inflammation, we explored whether the subsets and processes in resting intestine were altered in local inflammation.
At 4 days after inducing acute colitis by feeding 2% dextran sodium sulfate (DSS) to CX3CR1gfp/+ mice, there was intense infiltration of the mucosa by CD11b+ cells and this increased further by day 6 (Figure 4a). This was characterized by marked increases in the proportion and absolute number of CX3CR1int cells, together with accumulation of CX3CR1− eosinophils and neutrophils. Most of the expansion within the CX3CR1int compartment was among the P1 and P2 cells, whereas the numbers of cells in P3 increased less (Figure 4b–f). In contrast, the proportions and numbers of CX3CR1hi mφ decreased as colitis progressed, although this difference was not statistically significant. As in resting colon, all the cells in P1–P4 were CD64+, most became CD11c+, and F4/80 increased progressively (Figure 4b–d). Although the presumptive DC in P5 also expanded, this was much reduced compared with the other CX3CR1int populations (Figure 4b, e, and f). As in resting colon, P5 cells were all CD11chi and CD64− (Figure 4c and d).
Effects of inflammation on composition of colonic myeloid cell populations. (a) CX3CR1+/gfp mice were fed 2% dextran sodium sulfate in the drinking water for 7 days to induce colitis and the absolute numbers of CD11b+ LP leukocytes per colon assessed at d4 and d6, and in resting mice. (b) Representative expression of CD11b and CX3CR1-green fluorescent protein (GFP) by live-gated CD45+ colonic cells (upper panels), expression of Ly6C and MHCII by CD11b+ CX3CR1int colonic cells (middle panels), and expression of F4/80 and CD11c among CX3CR1int Ly6C− MHCII+ fraction (lower panels) in resting mice and on d4 and d6 of colitis. (c) Expression of Ly6C and CD64 by CX3CR1int CD11b+ cells in resting mice and on d4 of colitis. (d) Expression of CD11c and F4/80 by subsets P1–P5 on d4 of colitis. (e) Absolute numbers of CX3CR1-defined P1, P2, P3, P4, and P5 subsets of colonic CD11b+ cells during colitis. (f) Relative proportions of P1–P5 subsets among total CX3CR1+ cells in resting mice and on d4 and d6 of colitis. *P<0.05, **P<0.01, and ***P<0.001.
Thus, the same subsets of mφ are present in healthy and inflamed colon, but inflammation causes selective accumulation of cells phenotypically most similar to Ly6Chi monocytes.
Maturation of Ly6Chi monocytes is disrupted in intestinal inflammation
As these findings suggested that the local differentiation of Ly6Chi monocytes might differ between resting and inflamed colon, we examined this directly by transferring Ly6Chi monocytes into colitic recipients. As in resting mice, donor cells that had migrated to the colon at 24 h were found in the P1 and P2 populations (Figure 5a–c). By 96 h, 60% of the donor cells had acquired MHCII, upregulated F4/80, and the majority expressed intermediate or low levels of Ly6C. However, in contrast to what we found in “steady state” recipients, the donor monocytes did not become CX3CR1hi (Figure 5a and d). A small proportion of donor cells (1–2%) were found in the F4/80− CD11chicompartment of CX3CR1int cells (Figure 5b). As in resting mice, transferred Ly6Clo monocytes could be found in the bloodstream, but not the colon of mice with colitis (Supplementary Figure S5d online).
Effects of inflammation on recruitment of monocytes to colon. Monocyte recruitment into inflamed colon was studied by transferring 2 × 106 fluorescence-activated cell sorted Ly6Chi bone marrow monocytes from CD45.1+/CD45.2+ CX3CR1+/gfp mice into CD45.1+ wild-type (WT) mice on d3 of dextran sodium sulfate (DSS) colitis. After a further 2 days of DSS, mice were returned to normal drinking water. (a) Donor-derived cells within the live-gated CD45+ CD11b+ fraction of colonic LP cells of colitic mice 24 and 96 h after transfer compared with PBS-injected controls. CX3CR1int and CX3CR1hi gates were set using donor-derived monocytes in blood, which remain CX3CR1int. (b) Expression of F4/80 and CD11c by donor-derived cells in inflamed colon 24 and 96 h after transfer, compared with endogenous CD45+ CD11b+SSClo cells in colon on d4 DSS. (c) Expression of Ly6C and MHCII by F4/80+ donor-derived cells in inflamed colon 24 and 96 h after transfer, compared with endogenous CD45+ CD11b+ F4/80+ SSClo cells in colon on d4 DSS. (d) Mean fluorescence intensity (MFI) of CX3CR1-green fluorescent protein (GFP) expression on donor-derived monocytes derived from the colon and blood of resting CCR2−/− mice or from WT mice taken on day 4 of DSS colitis, with two mice per group. Results are representative of two individual experiments. PBS, phosphate-buffered saline.
Thus, inflammation appears to disrupt full differentiation of “inflammatory” Ly6Chi monocytes into CX3CR1hi resident mφ.
Mφ development in healthy colon is characterized by interleukin 10 production, but pro-inflammatory cells accumulate in inflammation
Next, we investigated the functional consequences of the differences in mφ found in steady-state and inflamed colon. Intracellular cytokine staining showed that all subsets from healthy colon could produce TNFα and IL10 under baseline culture conditions. A surprising number of cells produced both TNFα and IL10 in most populations (Figure 6a and b; Supplementary Figure S7a online). However, the proportion of cells producing IL10 alone increased progressively from P1 to P4. This was confirmed using Vert-X IL10 reporter mice (Supplementary Figure S7b online). Conversely there was a parallel, progressive decrease in the proportion of cells producing TNFα alone from P1 to P4 and few P1 cells made both IL10 and TNFα compared with the other subsets (Figure 6a and b). The progressive expression of IL10 from P1 to P4 was confirmed by PCR analysis. This also showed similar levels of TNFα mRNA in the different populations (Figure 6c).
Functional characterization and Toll-like receptor (TLR) responsiveness of colonic CX3CR1-defined myeloid cells in healthy and inflamed intestine. (a) Colonic LP cells isolated from resting CX3CR1+/gfp mice, or from mice receiving 2% dextran sodium sulfate (DSS) for 4 days, were cultured for 4.5 h in medium (top panels) or with 1 μg ml−1 lipopolysaccharide (LPS) (lower panels). Interleukin 10 (IL10) and tumor necrosis factor α (TNFα) production was assessed by intracellular cytokine staining. Results shown are mean proportions of IL10+, TNFα+, and IL10+TNFα+ cells within P1, P2, P3, and P4 cells in each condition + 1 s.d. for 3–4 mice per group. (b) Proportions of total cytokine-producing cells within P1 to P4 that produce IL10 alone, TNFα alone, or both IL10 and TNFα in resting (top panels) or inflamed colon (lower panels) ± lipopolysaccharide (LPS). Data representative of two individual experiments. (*P<0.05, **P<0.01, ***P<0.001 and ¶P<0.05, ¶¶P<0.01, ¶¶¶P<0.001 versus P1–P4 subsets from resting mice cultured in medium alone; †P<0.05, ††P<0.01, †††P<0.001 versus P1–P4 subsets from colitic mice cultured in medium alone.) (c) Quantitation of messenger RNA (mRNA) for IL10 and TNFα by fluorescence-activated cell sorted subsets P1–P4 from resting colon. (d) Quantitation of mRNA for pro-inflammatory cytokines and TLR by P1 subset of colonic myeloid cells from resting and colitic mice. Results are shown as mean expression relative to cyclophilin A (CPA) using the 2−ΔC(t) and are from three independent experiments using pooled cells from 12 mice per experiment (resting mice) or one experiment using pooled cells from 10 mice fed DSS for 4 days.
Significantly greater proportions of the CX3CR1int populations produced TNFα alone in inflamed colon compared with their counterparts in resting mice (Figure 6a and b). Mirroring their relative contributions to the inflammatory infiltrate, TNFα production was increased most markedly in P1 and P2. Very few P1 cells produced IL10 either alone or together with TNFα, and although the cells in P2 and P3 showed some shift toward IL10 production as they did in resting colon, most of this was accounted for by cells producing both TNFα and IL10 (Figure 6a and b). In contrast, CX3CR1hi mφ (P4) from colitic mice showed no increase in cells producing TNFα, either alone or in combination with IL10. Indeed there was a small, but significant, increase in IL10-producing cells among P4 in colitis compared with resting mice (Figure 6a and b).
Quantitative reverse transcription PCR analysis showed that pro-inflammatory gene expression was restricted to cells in P1, which showed increased expression of IL6, IL1β, TLR2, and TLR4 transcripts compared with resting P1 cells (Figure 6d). In contrast, mRNA for these genes did not change in the other subsets during colitis and the expression of other, non-inflammatory genes was not affected by inflammation in any population (Supplementary Figure S8 online).
Resistance to TLR stimulation develops as monocytes mature in healthy colon, but responsive cells accumulate in inflammation
To test the functional capacity of the subsets in resting and inflamed intestine further, we compared their responses to TLR stimulation. Lipopolysaccharide-stimulated P1 and P2 cells from normal colon showed robust increases in TNFα production as assessed by intracellular cytokine staining (Figure 6a and b). Although a substantial proportion of stimulated P1 or P2 cells also produced IL10, this was mostly in combination with TNFα and isolated production of IL10 decreased. In contrast, P3 cells showed no increases in cytokine production in response to lipopolysaccharide stimulation, while CX3CR1hi mφ showed only a small increase in IL10/TNFα dual-producing cells (Figure 6a and b). Stimulation with the TLR2 ligand BLP revealed a similar pattern of acquired TLR hyporesponsiveness (data not shown). This was not owing to loss of TLR expression, as all the phenotypic subsets expressed TLR1-9 mRNA at levels equivalent to or greater than those of BM-derived mφ (Supplementary Figure S9a online), as well as similar levels of TLR2 and CD14 protein (Supplementary Figure S9b online).
Lipopolysaccharide stimulation increased TNFα-producing cells within most colitic populations compared with unstimulated cells. However, isolated TNFα production was limited mostly to P1 and P2 cells, and it diminished progressively as cells matured. TLR4 stimulation caused a loss of cells producing IL10 alone in most populations (Figure 6a and b). A similar pattern of responsiveness was seen when colitic cells were stimulated with BLP (data not shown).
Thus, there is progressive differentiation of Ly6Chi monocytes toward an IL10-producing, TLR hyporesponsive population of resident mφ in steady-state colon. This process appears to be disrupted in colitis, with accumulation of pro-inflammatory CX3CR1int cells.
Context-dependent differentiation of mφs occurs in resting and inflamed human intestine
We next determined whether analogous processes occurred in human intestine. Monocytes and mφ were defined in resected ileal mucosa from healthy subjects as CD14+ Lin− cells and the majority of these were CD14lo (Figure 7a), consistent with them being resident mφ.16, 17 The majority of these CD14lo cells co-expressed HLA-DR with CD209 and/or CD163, but did not express CD11c, a marker found on circulating “inflammatory” monocytes in man.18 These CD14lo mφ therefore appear to be equivalent to CX3CR1hi mφ in mouse colon. Some of the MHCII+CD163lo cells within the CD14lo population did express CD11c, suggesting they may be similar to the later transitional stages of CX3CR1int cells in mice. The CD14hi population was more heterogeneous, comprising three subsets based on their expression of MHCII and CD163/CD209. These were HLA-DRloCD209loCD163lo, HLA-DRhiCD209loCD163lo, and HLA-DRhiCD209hiCD163hi (Figure 7a and b). This last subset had low levels of CD11c, suggesting these are relatively mature cells. These CD163+ mφ were located immediately below the epithelial surface and did not overlap with CD103+ DC (Figure 7c). In contrast, the other CD14hi cells were CD11chi and this was more marked on the HLA-DRloCD209loCD163lo cells, suggesting these are less mature than the other subsets. Thus, the CD14hi population of human mφ appears to comprise analogous differentiation intermediaries to those found among CX3CR1int mφ in mouse colon, with the HLA-DRloCD209loCD163lo, HLA-DRhiCD209loCD163lo, and HLA-DRhiCD209hiCD163hi subsets being homologous to P1, P2, and P3, respectively.
Macrophage subsets in the human ileum. Lin− (CD3, CD19, CD20, CD56, TCRαβ), live (PI−) singlet cells were identified in digests of resected healthy ileum (n=8) and CD45+ cells purified by magnetic antibody cell sorting. (a) CD14hi and CD14lo cells were identified and HLA-DR expression assessed. (b) Subsets of CD209/CD163− DR− (blue gate), CD209/CD163− DR+ (red gate), and CD209/CD163+ DR+ (green gate) cells were then identified among the CD14hi (top panels) and CD14lo cells (lower panels), and their expression of CD11c assessed. Plots are representative of eight (A) or three (B) individual samples. (c). Anatomical location of resident mφ in healthy human ileum. CD163+ mφ (green) are found immediately below the epithelial layer (46-diamidino-2-phenyl indole (DAPI)—blue) and are a discrete population from CD103+ DC and lymphocytes (red) (× 100–× 400 final magnification). (d) Representative plots identifying CD14hi and CD14lo CD103− cells among CD45+ Lin− cells from samples of healthy ileum, non-inflamed, or inflamed areas of Crohn's disease ileum (top panels). CD11c and CD163 expression were then assessed on the CD14hi subsets from each sample (lower panels). Results representative of 4–8 individual samples. (e) Ratios of CD14hi:CD14lo CD103− mφ in healthy, non-inflamed, and inflamed Crohn's disease ileal samples. Each symbol represents an individual patient. (f) CD64 vs. CD14 expression by live Lin− MHCII+ cells from normal and inflamed Crohn's disease human ileum. The left panels in each pair show CD64 isotype staining. ***P<0.001.
To determine whether the human and murine mφ subsets behaved similarly in inflammation, we analyzed human mφ in non-inflamed and inflamed ileum from patients with Crohn's disease (Figure 7d). While non-inflamed regions displayed a similar ratio of CD14hi:CD14lo mφ to that seen in healthy resected ileum, there was a dramatic accumulation of CD14hi mφ in inflamed tissue, to the extent that there was no longer a discrete population of CD14lo mφ (Figure 7d and e). As in healthy intestine, CD14hi mφ from non-inflamed tissue contained both CD163loCD11chi and CD163hiCD11clo subsets (Figure 7d). In marked contrast, the CD14hi population in inflamed Crohn's ileum was almost exclusively CD163lo CD11chi. Together, these results suggest there is accumulation of early monocyte-like cells in inflamed human intestine, and that the normal maturation of mφ may be disrupted during inflammation as it was in mice. As in mice, the CD14hi and CD14lo subsets of DR+ mφ were uniformly CD64+ in both healthy and inflamed human intestine, whereas MHCII+CD14− cells that include DC were mostly CD64− (Figure 7f).
Discussion
One of the most contentious challenges in mucosal immunology in recent years has been to identify the distinctive roles of MP populations such as mφ and DC, owing to reliance on markers that do not discriminate clearly between these cells.10 Here, we show that accurate identification of intestinal mφ and DC requires simultaneous assessment of CD11b, F4/80, CD11c, MHCII, CD64, Ly6C, and CX3CR1 expression by multiparameter flow cytometry. Using this rigorous approach, we demonstrate that the mφ pool in the mouse and human intestine is heterogeneous, with its exact nature being determined by the presence or absence of inflammation.
In resting mouse colon, most mφ express unusually high levels of CX3CR1, but there is also a significant population of CX3CR1int cells that expands preferentially in inflammation. Similar CX3CR1-based populations of intestinal mφ have been described recently,11 but we show for the first time that these populations are not independent subsets as previously assumed. Rather they represent stages in a single differentiation continuum from Ly6Chi monocytes to mature CX3CR1hi mφ. Our adoptive transfer, flow cytometry, and gene expression approaches all showed that Ly6Chi monocytes enter the colonic mucosa and differentiate in situ, with stepwise acquisition of MHCII, loss of Ly6C, and upregulation of F4/80, CD64, CD11c, and CX3CR1. There is also progressive expression of CD163, CD206, and TGFβR2, characteristic markers of resident intestinal mφ.17, 19 We show these phenotypic processes are accompanied by incremental changes in function, with incremental acquisition of phagocytic activity and IL10 production, loss of pro-inflammatory features such as IL6 and iNOS expression, and desensitization to TLR ligands. An identical “waterfall” of Ly6Chi monocytes developing progressively into CX3CR1hi class II MHChi CD64hi resident mφ in resting mouse intestine has also been proposed in a recent study conducted in parallel.20
The development of resident CX3CR1hi mφ in the intestine seems to be driven by the intestinal environment, as adoptively transferred Ly6Chi monocytes only became CX3CR1hiMHCIIhiF4/80hi in the mucosa and endogenous mφ with this phenotype are not seen in other tissues.21 Although Ly6Chi monocytes have been reported to give rise to “resident” mφ in atherosclerotic blood vessels,22 spinal cord,23 pregnant uterus,24 tumors,25 and skeletal muscle,26 all these models involved initial inflammation or disrupted tissue architecture. Notably, the progeny of Ly6Chi monocytes in the intestine did not express M2 mφ genes associated with tissue repair.19 Rather, they are more similar to “M2-like”/regulatory mφ found in tumors, where activation in the presence of factors such as IL10, TGFβ, or regulatory T cell extinguishes pro-inflammatory functions.19 Significantly, although some of these agents such as IL10 and TGFβ have also been implicated in regulating TLR signaling in intestinal mφ,17, 27, 28 it is unknown how they influence the development of the other phenotypic and functional properties of these cells. Similarly, while the presence of the local microbiota appears to be needed for production of IL10 by mucosal mφ,27, 29 conflicting findings have been reported on whether the composition of the mφ pool is altered in germ-free conditions.29, 30, 31 In our hands, there were also no differences in mφ subset proportions between the small and large intestines, despite vast differences in bacterial loads in these sites (our unpublished observations). The processes underlying intestinal mφ development are the subject of ongoing study.
We propose that similar processes may be taking place in normal human ileum. Consistent with other mature tissue mφ in man,19 most resident intestinal mφ were CD14loMHCII+ CD163/CD209hi, and as in mice6, 32 they were discrete from CD103+ DC, were found immediately below the surface epithelium and expressed CD64 preferentially. Transitional phenotypes analogous to those in mice could also be identified, starting with CD14hi CD11chi monocytes that appear to downregulate CD14, and acquire MHCII and CD163/CD209. Definitive proof that these phenotypic features do indeed reflect a differentiation continuum that is common to both species awaits establishment of methods for full functional analysis of the individual human subsets.
Although most previous work has concluded that pro-inflammatory mφ in the intestine are derived from a distinct population of monocytes,6 our study shows that inflammation is associated with disrupted differentiation of the same Ly6Chi monocytes that replenish resident mφ in resting intestine. As a result, monocytes and early stages of their differentiation continuum dominated the inflammatory infiltrate in DSS colitis. A similar phenomenon can be seen in T cell dependent colitis in mice20 and was also observed in inflamed Crohn's disease mucosa. Here there was accumulation of CD14hiCD11chi cells that appear to be the human equivalent of Ly6Chi monocytes in mice.33 In mice, the inflammatory cells in P1/P2 accounted for the upregulation of TNFα, IL1, and IL6 production, and hyper-responsiveness to TLR stimulation. Conversely, remaining CX3CR1hi mφ retained their bias toward IL10 production, confirming previous suggestions that resident mφ do not alter during inflammation.6 Accumulation of CX3CR1int mφ was reported previously in T-cell-dependent colitis in mice,31 but these authors did not detect monocytes in the mucosa and did not characterize the precursors of the CX3CR1int cells. The pathogenic role of newly recruited monocytes is supported by the fact that CCR2−/− mice fail to develop experimental colitis owing to defective recruitment of Ly6Chi monocytes.6 We are currently investigating the mechanisms underlying the failure of monocytes to differentiate fully during inflammation. It could be secondary to depletion of the factors that normally specify development into the resident mφ phenotype, perhaps because the destruction of mucosal architecture leads to loss of stromal and epithelial cells that produce trophic mediators. Alternatively, the pro-inflammatory mediators that accumulate in colitis may have direct inhibitory effects on monocyte development.
Although previous studies have shown that transferred Ly6Chi monocytes can migrate to the non-inflamed intestine, these used intense depletion of the resident mφ compartment and the mucosal progeny were often identified as a subset of DC based on expression of CD11c and MHCII.14 While our manuscript was in preparation, Rivollier et al.29 also reported recruitment of Ly6Chi monocytes to the inflamed colon, but concluded that the resulting cells were inflammatory “DC”. They proposed that monocyte differentiation is diverted selectively into the DC lineage under these conditions. In contrast, our results indicate that these inflammatory cells belong to the mφ lineage. They are F4/80+CD11c+MHCII+ and express the CD64 marker we have shown is not present on DC.12 Furthermore, they are phenotypically and functionally identical to the flt3L unresponsive P2 and P3 cells we found in resting mucosa. Distinguishing these lineages is important, because the mφ subsets have properties such as IL10 production and phagocytic activity not shared by bona fide mucosal DC, indicating these will have quite distinct roles in vivo. Thus, myeloid cells in the mucosa need to be defined precisely using a combination of functional and phenotypic criteria.
Our findings that resident intestinal mφ are replenished continuously by CCR2-dependent recruitment of Ly6Chi monocytes even in the absence of inflammation are consistent with the reduction in IL10-producing gut mφ seen in CCL2-deficient mice.34 Although we cannot exclude the possibility that some resident mφ may be derived from long-lived, self-renewing precursors, as seen in the brain or skin,32 we could find no evidence of cell division in situ among any of the intestinal mφ subsets. Importantly, the so-called “resident” subset of Ly6Clo monocytes also does not appear to contribute to the mφ in the resting or inflamed intestine. Continual replenishment by functionally plastic monocytes would be very appropriate for the intestinal immune system, where constant exposure to bacteria and other materials requires rapid and aggressive responses to potential pathogens. Together with their expression of scavenger receptors, high IL10 production, phagocytic activity, and anatomical position beside the epithelium, resident mφ are ideally suited for maintaining the symbiotic relationship between the host and its microbiota. They do this by acting as microbial waste disposal units without provoking tissue damage and by driving the secondary expansion of antigen-specific regulatory T cells.3, 4 The primary effect of inflammation is to disrupt the differentiation processes that normally produce these homeostatic mφ, although it is unclear whether this reflects loss of intrinsic factors that usually specify full maturation, or if inflammation actively revises these processes. Targeting the factors that drive the differentiation of monocytes under different circumstances may be more beneficial for treating IBD than blocking recruitment or development of individual monocyte populations.
Methods
Mice. Wild-type C57Bl/6 (B6) (Harlan Olac, Bicester, UK), CX3CR1+gfp,35 C57Bl/6. SJL (CD45.1+), CCR2−/−,36 CD11c-DTR-GFP, and IL10eGFP mice37 were maintained under specific pathogen-free conditions at the Central Research Facility at the University of Glasgow. All mice had been backcrossed for at least nine generations on to the B6 background and were used at 6–12 weeks of age.
Patients and tissues. Macroscopically inflamed and non-inflamed ileal tissue was obtained from biopsies (n=4) or surgical specimens (n=3) from Crohn's patients undergoing colonoscopy or ileocoecal resections, respectively (Supplementary Table S1 online). Their median age was 31 years (range, 23–38), and the median disease duration was 6 years. Biopsy samples were obtained from patients that had no current medication. Resection samples of healthy terminal ileum were obtained from patients (median age 68, range 59–78), 6 male, 2 female) undergoing cystectomy with intestinal neobladder reconstruction (n=8).
Isolation of lamina propria cells. Lamina propria cells were obtained from mouse colon by enzymatic digestion as described previously.6, 38 For morphological assessment, cells were spun onto Polysine glass microscope slides (VWR International, East Grinstead, UK), fixed in acetone, and stained using the Rapid-Romanowsky staining kit (Raymond A. Lamb, Eastbourne, UK).
For surgical specimens of human ileum, the muscle, and fat were removed and the remaining tissue was cut into small (<5 mm) pieces. Tissue fragments were incubated in Hank's balanced salt solution containing ethylenediaminetetraacetic acid (2 mm) for 3 × 15 min at 37 °C, and after each incubation, epithelial cells were removed by passage through a nylon filter as previously described.39 Human ileal biopsies were incubated in Hank's balanced salt solution/ethylenediaminetetraacetic acid for 45 min at 37 °C with rotation (250 rpm) and the cells in suspension were discarded. Remaining tissue fragments were digested for 2 × 60 min at 37 °C in R10 medium (RPMI, 10% FCS, 10 mm HEPES, 100 U ml−1 penicillin, 100 μg ml−1 streptomycin, and 50 μg ml−1 gentamycin (all Gibco, Invitrogen, Paisley, UK)) containing 0.025 Wunch U ml−−1 Liberase (Roche Diagnostics GmbH, Mannheim, Germany)/20 IU ml−1 DNAse I (Sigma, Poole, UK) with magnetic stirring. Digested material was filtered through a 100-μm cell strainer and pooled before analysis.
Fluorescence microscopy. Endogenous peroxidase activity of acetone-fixed human ileal cryosections (7 μm) was blocked by incubating sections for 10 min with 0.5% H2O2. Slides were then incubated with donkey serum (10%) in phosphate-buffered saline-Tween20 (0.05%) (Sigma) and avidin–biotin blocking kit according to manufacturer's instructions (Vector Laboratories, Burlingame, CA). Tissue was then stained with mouse anti-human CD163 (1 μg ml−1, BerMac3, DAKO, Cambridge, UK) for 45 min followed by Alexa-488-labeled donkey anti-mouse antibody (Jackson ImmunoResearch Laboratories, West Grove, PA). After washing and blocking with 10% mouse serum for 30 min, sections were stained with biotinylated CD103 (1 μg ml−1) (28C12, kind gift from Dr M. Brenner, Brigham and Women's Hospital, Boston, MA) in the presence of 10% mouse serum for 45 min. Slides were visualized using a biotin-tyramide signal amplification kit according to the manufacturer's recommendations (PerkinElmer Life Science, Cambridge, UK) with Alexa-555-conjugated streptavidin and 46-diamidino-2-phenyl indole. Isotype control antibodies were included in each staining as controls. Images were acquired with an Axiovert 200 m microscope (Carl Zeiss MicroImaging, Jena, Germany) and Volocity 5.2.1 software (Improvision Perkin Elmer, Coventry, UK).
Flow cytometric analysis and sorting of cells. 1–5 × 106 cells were stained at 4 °C in the dark as described previously38 using the antibodies listed in Supplementary Table S2 online and analyzed using an LSR II or FACSAria I (BD Biosciences, Oxford, UK) and FlowJo software (Tree Star, Ashland, OR). To detect intracellular cytokines, cells were incubated in complete RPMI at 37 °C in 5% CO2 for 4.5 h in the presence of 1 μm monensin and 10 μg ml−1 Brefeldin A (both Sigma) in 12 × 75 mm polystyrene tubes (BD Falcon, Oxford, UK). After cell surface staining, cells were fixed with 4% paraformaldehyde (Thermo Scientific, Cramlington, UK) at room temperature for 10 min, washed in phosphate-buffered saline and permeabilized using Cytoxfix/Cytoperm (BD Biosciences). After a further incubation with purified anti-CD16/CD32, cells were stained with anti-TNFα-APC and anti-IL10-PE (both BD Biosciences) or isotype controls for 20 min, washed, and analyzed.
Flt3L-mediated expansion of cells in vivo. CX3CR1+/gfp mice were injected intraperitoneally with 10 μg human recombinant CHO-derived Flt3L (a kind gift of Amgen, Seattle, WA) in 0.2 ml sterile phosphate-buffered saline for eight consecutive days.
Adoptive transfer of bone marrow monocytes. Bone marrow cells from CX3CR1+/gfp CD45.2+ or CX3CR1+/gfp CD45.1+/CD45. 2+ mice were stained for CD11b-AF700, CD117-APC-H7, Ly6G-PE, and Ly6C-PE-Cy7. Ly6Chi monocytes (CD11b+CD117−Ly6G−Ly6ChiCX3CR1int) and Ly6Clo monocytes (CD11b+ CD117−Ly6G−Ly6Clo CX3CR1+) were sorted to >97% purity using a FACSAria I cell sorter (Supplementary Figure S3a online). CD11c-DTR recipient mice received 4 ng g−1 bodyweight diphtheria toxin (Sigma) 24 h before monocyte transfer.
Assessment of BrdU incorporation in vivo. Mice were injected intraperitoneally with 1 mg BrdU (BD Biosciences) and the incorporation of BrdU by isolated cells was assessed using the BD BrdU Flow Kit (BD Biosciences).
Induction of DSS colitis. Mice received 2% DSS salt (reagent grade; MW 36,000–50,000 kDa; MP Biomedicals, Solon, OH) ad libitum in sterile drinking water for up to 8 days as described previously.6, 38
Assessment of phagocytosis. 3 × 106 cells were assessed for phagocytosis of pHrodo Escherichia coli bioparticles (Life Technologies, Paisley, UK) according to the manufacturer's guidelines and analyzed by flow cytometry.
Quantitation of gene expression by real-time reverse transcription PCR. Total RNA was purified from sorted CX3CR1-defined LP cells using the RNeasy Micro kit (Qiagen, Crawley, UK). Thirteen nanograms RNA was reverse transcribed to complementary DNA using the Superscript II First strand synthesis system (Invitrogen, Paisley, UK). Gene expression was assayed by quantitative reverse transcription PCR using Brilliant III Ultra Fast SYBR qPCR master mix (Agilient Technologies, Workingham, UK) on the 7500HT Fast system (Applied Biosystems, Paisley, UK). Primers (Integrated DNA Technologies, Glasgow, UK) as detailed in Supplementary Table S3 online. Complementary DNA samples were assayed in triplicate and gene expression levels were normalized to Cyclophilin A. The mean relative gene expression was calculated using the 2−ΔC(t) method.
Statistical analysis. Results are presented as means±1 s.d., and groups were compared using a Student's t-test, Mann–Whitney test, or for multiple groups, a one-way ANOVA followed by a Bonferroni post test using Prism Software (GraphPad Software, La Jolla, CA).
References
Bain, C.C. & Mowat, A.McI. Intestinal macrophages - specialised adaptation to a unique environment. Eur. J. Immunol. 41, 2494–2498 (2011).
Geissmann, F., Manz, M.G., Jung, S., Sieweke, M.H., Merad, M. & Ley, K. Development of monocytes, macrophages, and dendritic cells. Science 327, 656–661 (2010).
Hadis, U. et al. Intestinal tolerance requires gut homing and expansion of foxP3+ regulatory T cells in the lamina propria. Immunity 34, 237–246 (2011).
Kayama, H. et al. Intestinal CX3C chemokine receptor 1high (CX3CR1high) myeloid cells prevent T-cell-dependent colitis. Proc. Natl Acad. Sci. USA 109, 5010–5015 (2012).
MacDonald, T.T., Monteleone, I., Fantini, M.C. & Monteleone, G. Regulation of homeostasis and inflammation in the intestine. Gastroenterology 140, 1768–1775 (2011).
Platt, A.M., Bain, C.C., Bordon, Y., Sester, D.P. & Mowat, A.McI. An independent subset of TLR expressing CCR2-dependent macrophages promotes colonic inflammation. J. Immunol. 184, 6843–6854 (2010).
Geissmann, F., Gordon, S., Hume, D.A., Mowat, A.McI. & Randolph, G.J. Unravelling mononuclear phagocyte heterogeneity. Nat. Rev. Immunol. 10, 453–460 (2010).
Bogunovic, M. et al. Origin of the lamina propria dendritic cell network. Immunity 31, 513–525 (2009).
Schulz, O. et al. Intestinal CD103+, but not CX3CR1+, antigen sampling cells migrate in lymph and serve classical dendritic cell functions. J. Exp. Med. 206, 3101–3114 (2009).
Persson, E.K., Jaensson, E. & Agace, W.W. The diverse ontogeny and function of murine small intestinal dendritic cell/macrophage subsets. Immunobiology 215, 692–697 (2010).
Weber, B., Saurer, L., Schenk, M., Dickgreber, N. & Mueller, C. CX3CR1 defines functionally distinct intestinal mononuclear phagocyte subsets which maintain their respective functions during homeostatic and inflammatory conditions. Eur. J. Immunol. 41, 773–779 (2011).
Langlet, C. et al. CD64 expression distinguishes monocyte-derived and conventional dendritic cells and reveals their distinct role during intramuscular immunization. J. Immunol. 188, 1751–1760 (2012).
Gratchev, A. et al. Activation of a TGF-β-specific multistep gene expression program in mature macrophages requires glucocorticoid-mediated surface expression of TGF-beta receptor II. J. Immunol. 180, 6553–6565 (2008).
Varol, C. et al. Intestinal lamina propria dendritic cell subsets have different origin and functions. Immunity 31, 502–512 (2009).
Varol, C. et al. Monocytes give rise to mucosal, but not splenic, conventional dendritic cells. J. Exp. Med. 204, 171–180 (2007).
Smith, P.D. et al. Intestinal macrophages lack CD14 and CD89 and consequently are down-regulated for LPS- and IgA-mediated activities. J. Immunol. 167, 2651–2656 (2001).
Smith, P.D., Smythies, L.E., Shen, R., Greenwell-Wild, T., Gliozzi, M. & Wahl, S.M. Intestinal macrophages and response to microbial encroachment. Mucosal Immunol. 4, 31–42 (2011).
Saha, P. & Geissmann, F. Toward a functional characterization of blood monocytes. Immunol. Cell. Biol. 89, 2–4 (2011).
Sica, A. & Mantovani, A. Macrophage plasticity and polarization: in vivo veritas. J. Clin. Invest. 122, 787–795 (2012).
Tamoutounour, S. et al. CD64 distinguishes macrophages from dendritic cells in the intestine and reveals the Th1-inducing role of mesenteric lymph node macrophages during colitis. Eur. J. Immunol. advance online publication, 31 August 2012; doi: 10.1002/eji.201242847 (e-pub ahead of print).
Winnall, W.R., Muir, J.A. & Hedger, M.P. Rat resident testicular macrophages have an alternatively activated phenotype and constitutively produce interleukin-10 in vitro. J. Leukoc. Biol. 90, 133–143 (2011).
Tacke, F. et al. Monocyte subsets differentially employ CCR2, CCR5, and CX3CR1 to accumulate within atherosclerotic plaques. J. Clin. Invest. 117, 185–194 (2007).
Shechter, R. et al. Infiltrating blood-derived macrophages are vital cells playing an anti-inflammatory role in recovery from spinal cord injury in mice. PLoS Med. 6, e1000113 (2009).
Svensson, J., Jenmalm, M.C., Matussek, A., Geffers, R., Berg, G. & Ernerudh, J. Macrophages at the fetal-maternal interface express markers of alternative activation and are induced by M-CSF and IL-10. J. Immunol. 187, 3671–3682 (2011).
Movahedi, K. et al. Different tumor microenvironments contain functionally distinct subsets of macrophages derived from Ly6Chigh monocytes. Cancer Res. 70, 5728–5739 (2010).
Arnold, L. et al. Inflammatory monocytes recruited after skeletal muscle injury switch into antiinflammatory macrophages to support myogenesis. J. Exp. Med. 204, 1057–1069 (2007).
Ueda, Y. et al. Commensal microbiota induce LPS hyporesponsiveness in colonic macrophages via the production of IL-10. Int. Immunol. 22, 953–962 (2010).
Smythies, L.E. et al. Inflammation anergy in human intestinal macrophages is due to Smad-induced IκBα expression and NF-κB inactivation. J. Biol. Chem. 285, 19593–19604 (2010).
Rivollier, A., He, J., Kole, A., Valatas, V. & Kelsall, B.L. Inflammation switches the differentiation program of Ly6Chi monocytes from antiinflammatory macrophages to inflammatory dendritic cells in the colon. J. Exp. Med. 209, 139–155 (2012).
Haverson, K., Rehakova, Z., Sinkora, J., Sver, L. & Bailey, M. Immune development in jejunal mucosa after colonization with selected commensal gut bacteria: a study in germ-free pigs. Vet. Immunol. Immunopathol. 119, 243–253 (2007).
Niess, J.H. & Adler, G. Enteric flora expands gut lamina propria CX3CR1+ dendritic cells supporting inflammatory immune responses under normal and inflammatory conditions. J. Immunol. 184, 2026–2037 (2010).
Schulz, C. et al. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science 336, 86–90 (2012).
Rugtveit, J., Nilsen, E.M., Bakka, A., Carlsen, H., Brandtzaeg, P. & Scott, H. Cytokine profiles differ in newly recruited and resident subsets of mucosal macrophages from inflammatory bowel disease. Gastroenterology 112, 1493–1505 (1997).
Takada, Y. et al. Monocyte chemoattractant protein-1 contributes to gut homeostasis and intestinal inflammation by composition of IL-10-producing regulatory macrophage subset. J. Immunol. 184, 2671–2676 (2010).
Jung, S. et al. Analysis of fractalkine receptor CX(3)CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol. Cell. Biol. 20, 4106–4114 (2000).
Kurihara, T., Warr, G., Loy, J. & Bravo, R. Defects in macrophage recruitment and host defense in mice lacking the CCR2 chemokine receptor. J. Exp. Med. 186, 1757–1762 (1997).
Madan, R. et al. Nonredundant roles for B cell-derived IL-10 in immune counter-regulation. J. Immunol. 183, 2312–2320 (2009).
Bain, C.C. & Mowat, A.McI. CD200 receptor and macrophage function in the intestine. Immunobiology 217, 643–651 (2012).
Johansson-Lindbom, B. et al. Functional specialization of gut CD103+ dendritic cells in the regulation of tissue-selective T cell homing. J. Exp. Med. 202, 1063–1073 (2005).
Acknowledgements
We would like to thank Drs B. Jeppson, J. Marsal, U. Håkansson, G. Baseckas, and O. Patchan (Skåne University Hospital, Lund/Malmö, Sweden) for collecting human ileum. The authors are grateful to Drs Oliver Pabst and Vuk Cerovic for constructive review of the manuscript. CCB, CLS, and AMcIM were supported by the Wellcome Trust and MRC UK.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declared no conflict of interest.
Additional information
SUPPLEMENTARY MATERIAL is linked to the online version of the paper
Supplementary information
Rights and permissions
This work is licensed under the Creative Commons Attribution-NonCommercial-No Derivative Works 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Bain, C., Scott, C., Uronen-Hansson, H. et al. Resident and pro-inflammatory macrophages in the colon represent alternative context-dependent fates of the same Ly6Chi monocyte precursors. Mucosal Immunol 6, 498–510 (2013). https://doi.org/10.1038/mi.2012.89
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/mi.2012.89
This article is cited by
-
IL-10 constrains sphingolipid metabolism to limit inflammation
Nature (2024)
-
Cellular crosstalk in atherosclerotic plaque microenvironment
Cell Communication and Signaling (2023)
-
Metabolic heterogeneity of tissue-resident macrophages in homeostasis and during helminth infection
Nature Communications (2023)
-
MANF ameliorates DSS-induced mouse colitis via restricting Ly6ChiCX3CR1int macrophage transformation and suppressing CHOP-BATF2 signaling pathway
Acta Pharmacologica Sinica (2023)
-
CD301b+ macrophage: the new booster for activating bone regeneration in periodontitis treatment
International Journal of Oral Science (2023)