Abstract
Active fungal proteinases are powerful allergens that induce experimental allergic lung disease strongly resembling atopic asthma, but the precise relationship between proteinases and asthma remains unknown. Here, we analyzed dust collected from the homes of asthmatic children for the presence and sources of active proteinases to further explore the relationship between active proteinases, atopy, and asthma. Active proteinases were present in all houses and many were derived from fungi, especially Aspergillus niger. Proteinase-active dust extracts were alone insufficient to initiate asthma-like disease in mice, but conidia of A. niger readily established a contained airway mucosal infection, allergic lung disease, and atopy to an innocuous bystander antigen. Proteinase produced by A. niger enhanced fungal clearance from lung and was required for robust allergic disease. Interleukin 13 (IL-13) and IL-5 were required for optimal clearance of lung fungal infection and eosinophils showed potent anti-fungal activity in vitro. Thus, asthma and atopy may both represent a protective response against contained airway infection due to ubiquitous proteinase-producing fungi.
Similar content being viewed by others
Introduction
Atopic asthma has increased in prevalence and severity worldwide over the past 30 years, transforming this respiratory disorder into an epidemic ailment affecting much of the industrialized world.1, 2 Genetic factors clearly contribute to asthma susceptibility, but the slow rate of genetic change within large populations is unlikely to explain the recent increase in disease prevalence and severity. A more plausible explanation of the change in asthma epidemiology would invoke one or more environmental factors, alterations of which may lead to disease in genetically susceptible individuals.
Type 2T-helper (Th2) cell-mediated allergic inflammation is both a common finding in patients with diverse clinical asthma phenotypes and a critical determinant of experimental allergic lung disease.3, 4, 5, 6, 7 Major clinical signs and symptoms of asthma and asthma-related mortality are related to airway obstruction,8 which consists of at least three components: airway hyperresponsiveness (AHR), an exaggerated constrictive response to cholinergic agonists; physical obstruction of the airways by metaplastic airway epithelial goblet cells that secrete glycoproteins; and airway inflammation characterized by the presence of eosinophils. Experimental models have previously showed that allergic airway inflammation and airway obstruction are coordinated by Th2-related cytokines, especially interleukin 4 (IL-4), IL-5, IL-13, and IL-25.9, 10, 11, 12, 13
The lungs are remarkably tolerant of inhaled antigens. In experimental systems, antigens such as ovalbumin that are devoid of known adjuvant substances typically elicit T-regulatory responses that suppress active inflammation.14, 15 Thus, a paradox of allergic asthma is that despite the tolerogenic predisposition of the lung, asthma patients show lung and systemic inflammation and immune reactivity to a broad range of environmental antigens, termed atopy.16, 17, 18 This suggests the existence in human environments of adjuvant substances capable of bypassing lung tolerogenic mechanisms and establishing allergic inflammation in response to diverse inhaled agents.
As established through studies of mice, active proteinases are powerful initiators of allergic lung disease and are thus candidate adjuvant factors potentially underlying diseases such as asthma.14, 19 Experimentally, allergenic proteinases derive from diverse sources, but fungal proteinases are particularly potent inducers of the full spectrum of allergic lung disease.14, 20, 21 Moreover, proteinase-producing fungi are ubiquitous in human environments and have previously been strongly linked to human and experimental allergic lung disease.22, 23, 24, 25 However, although the link between environmental fungi and asthma is thought to be based on hypersensitivity to fungal products,26, 27 this important relationship has not been fully characterized.
In this study, we examined dust collected from the homes of asthmatic children in the Houston, Texas area to determine the presence and source(s) of active, potentially allergenic proteinases. We show the widespread existence of household dust proteinase activity and that common fungi are frequent sources of active household proteinases. More importantly, proteinase-producing fungi readily infected the mouse airway, producing both proteinase-dependent allergic lung disease and overcoming lung tolerogenic mechanisms to induce allergic responses to normally innocuous inhaled antigens. Although often believed to contribute to airway obstruction in asthma, eosinophils were found to be protective against fungus-induced allergic lung disease, suggesting that asthma may represent a protective response to contain at the mucosal surface airway infection by proteinase-producing fungi.
Results
Fungal, but not mite, proteinases are active in dust from asthmatic households
Proteinases derived from a variety of allergenic organisms (dust mites, fungi, pollens) are highly allergenic, but require intact proteinase activity.14 To determine if enzymatically active, potentially allergenic proteinases can be detected from homes, we analyzed 84 dust samples collected from 31 asthmatic households. These analyses revealed that significant quantities of total proteinase activity were present in 95% of dust samples (Table 1). Gelatin gel zymography further revealed proteinase bands of very high molecular size (60–220 kDa), exceeding the predicted size of most organismal proteinases (Figure 1).
House dust contains active fungal proteinases. Zymography was used to reveal active proteinases present in dust samples obtained from three representative houses and to determine their likely sources. The left lane of each pair depicts proteinases, visible as lucent bands, present in aqueous extracts of each dust sample. Right lanes of each pair depict proteinases secreted into culture medium containing the dominant fungus isolated from the corresponding dust sample. Dust extracts 1 and 3 show the common 86 kDa proteinase band present in over 50% of all samples; the fungus isolated from these samples was A. niger. Neither the dust nor the culture medium of the dominant fungus isolated from sample no. 2 shows proteinase activity.
Immunological analysis of dust samples revealed large quantities of the dust mite proteinase Der p 1 in 40% of dust samples (Table 1). Unexpectedly, however, careful review of all dust sample zymograms failed to reveal a 25 kDa proteinase band corresponding to the native molecular size of Der p 1 and similar in size to many other mite proteinases. In contrast, commercially available, enzymatically active Der p 1 was detected at its expected molecular size even when added to house dust samples (Supplementary Figure 1 online). Incubation of house dust samples under reducing conditions further failed to retrieve significant Der p 1 proteinase activity (Supplementary Figure 2 online). These findings indicate that Der p 1 that is immunologically detectable in dust samples does not have significant proteinase activity and is unlikely to account for the proteinase activity present in house dust samples.
To further investigate the potential sources of house dust proteinases, dust samples were analyzed by zymography to confirm the presence of discrete proteinases and to determine their molecular size. For selected extracts, the extracts were paired in zymograms with culture medium in which the dominant fungus from the same sample was grown. Representative examples of this analysis are shown in Figure 1. Based on matching molecular size of ∼86 kDa, apparent identity between a dust proteinase and the secreted fungal proteinase was established in two of three examples shown (Figure 1). The fungal proteinase was produced by Aspergillus niger in samples 1 and 3 and an apparently identical ∼86 kDa proteinase was present in half of all dust samples analyzed by this method (18/36; Table 1). Although this technique is not definitive, these findings are significant because our ongoing studies indicate that A. niger is ubiquitous (100%) in asthmatic households in the Houston area, suggesting that this fungus, and not dust mite species, is a major source of house dust proteinases.
A. niger conidia isolated from household dust induce robust allergic lung disease
The conidia of A. niger were devoid of proteinase activity (0 proteinase units (PU) mg–1 conidia where 1 PU=activity of 1 ng of A. oryzae aspergillopepsin (Sigma Chemical Co., St Louis, MO, p-4032)) whereas the hyphae of A. niger were substantially proteolytically active (3.82±0.02 PU mg–1 hyphae). Thus, if household hyphal-derived proteinases contribute to allergic lung disease, they could do so directly by eliciting allergic inflammation following inhalation. This possibility was remote because the proteinase activity of house dust samples far exceeding a physiologically reasonable inhaled burden (1 g) represented <0.1% of the minimal proteinase activity required to induce allergic lung disease in mice (∼7 PU vs. ∼7000 PU).14 Moreover, mice intranasally challenged over 2 weeks with an aqueous extract of house dust containing significant proteinase activity showed exclusively a neutrophil-predominant inflammatory response devoid of allergic features and no airway hyperresponsiveness (data not shown).
Alternatively, we reasoned that the conidia of allergenic fungi might produce active airway infection if inhaled and the allergenic proteinases produced continuously in situ might more efficiently induce allergic disease. To test this, we challenged mice with 400 × 103 viable or γ-radiation-sterilized A. niger conidia every 2 days for 16 days and assessed mice for the induction of allergic lung disease. At multiple acetylcholine (Ach) doses, viable A. niger conidia-challenged mice showed airway hyperresponsiveness as determined by enhanced Ach-induced increases in respiratory system resistance (RRS; Figure 2a).
A. niger conidia induce allergic lung disease. C57Bl/6 mice received intranasally 400 × 103 viable A. niger conidia (400 k), 400 × 103 non-viable irradiated A. niger conidia (400 k Irrad.) or PBS as indicated every other day for a total of eight challenges. Data were collected 24 h after the final challenge. (a) Airway hyperresponsiveness as assessed by the increase in respiratory system resistance (RRS) with increasing concentrations of Ach injected intravenously. (b) Total secreted glycoproteins quantitated from BAL fluid. (c) Total numbers of eosinophils (Eos), macrophages (Mac), neutrophils (Neu) and lymphocytes (Lymph) isolated from BAL fluid. (d) ELISpot-based quantitation of total IL-4- and IFN-γ-secreting cells from whole lung. (e) Concentration of indicated cytokines and chemokines as assessed by multiplex assay from bronchoalveolar lavage fluid. Data are from two experiments with n=3 or 4 animals. *P<0.05 relative to PBS-challenged animals; **P<0.05 comparing viable and irradiated conidia-challenged animals. BAL, bronchoalveolar lavage; PBS, phosphate-buffered saline.
Mice challenged with viable conidia also had markedly increased total secreted glycoproteins measured in bronchoalveolar lavage (BAL) fluid (Figure 2b), increased total BAL fluid cellularity, consisting largely of eosinophils (Figure 2c), and a predominance of IL-4 secreting cells in whole lung as assessed by ELISpot technique (Figure 2d; IL-4/IFN-γ ratio >100:1). Comprehensive analysis of bronchoalveolar lavage fluid cytokines confirmed the induction of predominant Th2-cell-derived cytokines (IL-4, IL-6, IL-13) in response to viable conidia challenge (Figure 2e). Pronounced peribronchovascular, eosinophil-predominant inflammation and goblet cell metaplasia were observed in hematoxylin and eosin- and PAS-stained sections of lung from mice challenged with live conidia (Figure 3a and b). Although rare, apparent fungal hyphae were observed in lung sections and were isolated to airway lumina (Figure 3c), whereas conidia were common and seen predominantly in lung interstitium (Figure 3d). Histological examination of bronchoalveolar lavage fluid confirmed the presence of fungal hyphae (Figure 3e and f).
A. niger conidia induce allergic lung inflammation and active fungal infection. C57Bl/6 mice were challenged with A. niger conidia as in Figure 2 and lungs were removed 24 h after the final challenge, fixed with formalin and prepared for hematoxylin and eosin (H&E; a) and periodic acid-Schiff (b) staining of 5 μm sections. Arrows indicate metaplastic goblet cells. (c,d) Gomori methenamine silver (GMS) staining of lung parenchyma reveals rare elongated airway luminal hyphae (c) and abundant spherical lung parenchymal conidia (d). (e,f) H&E staining of BAL fluid specimens from conidia-challenged mice showing small, septate hyphae apparently emerging from a conidium (arrow; e) with many adherent eosinophils (E) and macrophages (M) and a much larger mature hypha (f). BAL, bronchoalveolar lavage.
In contrast to viable conidia, sterile, irradiated conidia failed to induce airway hyperresponsiveness (Figure 2a). Glycoprotein secretion and airway eosinophilia were largely preserved or enhanced in mice receiving irradiated conidia, whereas comparatively greater numbers of macrophages and neutrophils were observed in BAL fluid (Figure 2b and c). Many fewer IL-4-secreting cells were recruited to lungs of mice receiving irradiated conidia and lung IFN-γ-secreting cells were significantly enhanced (Figure 2d; IL-4/IFN-γ ratio 1.6:1). Irradiated conidia further induced markedly less IL-4 and no IL-13 compared with viable conidia; moreover, non-viable conidia induced substantially greater secretion of the pro-inflammatory cytokines IL-17A and tumor necrosis factor and the neutrophil chemokine KC (Figure 2e). A. niger-specific IgG1 was detected in the sera of 85% of viable conidia-challenged animals, but fungus-specific IgE was not found (data not shown). Thus, a vigorous type 2 lung immune response associated with robust allergic lung disease was observed following challenge with viable A. niger conidia. In contrast, sterilized conidia induced predominant type 1 and type 17 lung immunity that failed to elicit a complete allergic lung disease phenotype.
Conidia of A. niger produce an active, contained infection following airway challenge
The preceding studies indicated that active fungal infection, and not conidia hypersensitivity, was required for allergic lung disease. To confirm this, we first determined the persistence of A. niger in lungs based on the ability to re-isolate the organism at increasingly distant time points following a single intranasal challenge. After inhalation of 400 × 103 conidia once, maximal airway eosinophilia was observed on the second day following challenge, but eosinophils persisted in bronchoalveolar lavage fluid for 1 week (Figure 4a). Moreover, viable organisms were recovered from bronchoalveolar lavage fluid for up to 7 days, with log-linear clearance kinetics (Figure 4b). These findings were suggestive of the persistence and perhaps growth of A. niger in the airways.
Persistence of A. niger infection in mouse airways. Mice received one intranasal challenge with 400 × 103 A. niger conidia. Thirty minutes and 1, 2, 5, and 7 days later (a) the percent abundance of inflammatory cells was determined from BAL fluid (BALF) and (b) fungal colony forming units (CFU) from lung and BAL fluid were determined and expressed as a percentage of the 30-min time point. Data are from one of two comparable experiments; n=3. BAL, bronchoalveolar lavage.
We performed additional dose-titration experiments to determine the minimum number of conidia required to establish a complete asthma phenotype. For these experiments, to mimic the presumed exposure patterns of humans to ubiquitous environmental fungi, mice were challenged daily with A. niger conidia. Very low dose challenge with conidia (1–2 × 103 conidia per dose) was sufficient to elicit some aspects of allergic lung disease, but a minimum dose of 50 × 103 conidia dose per day was required to elicit a complete allergic lung disease phenotype that included airway hyperresponsiveness (Figure 5a–e). Moreover, serum IgG1 reactivity to A. niger antigen was strongly dependent on the fungal infectious burden, with little or no reactivity seen with doses under 10 × 103 conidia per dose and 100% reactivity was observed only at the relatively high dose of 200 × 103 conidia per dose (Figure 5f).
Dose-dependent induction of allergic lung disease and IgG1 responses with A. niger infection. Mice received intranasally PBS, 1 × 103 (1 k), 2, 5, 10, 50, or 200 k viable A. niger conidia daily for 18 days and airway hyperresponsiveness (a,b), total BAL fluid (BALF) glycoproteins (c), total BAL fluid inflammatory cells (d) and total lung IL-4 and IFN-γ-secreting cells (e) were quantitated as in Figure 2. (f) The percent of the same mice with IgG1 reactivity to A. niger antigen as assessed by western blot. Data are from one of two comparable experiments, n=3 mice/group. Panel a *P<0.05 50 k vs. PBS; **P<0.05 200 k vs. PBS Panels b–e *P<0.05 relative to PBS. BAL, bronchoalveolar lavage.
As with much larger numbers of conidia administered discontinuously (Figure 2), lethal irradiation of A. niger conidia abolished the ability of daily exposure to 50 × 103 conidia to induce marked lung IL-4 responses and airway hyperreactivity, although again substantial airway eosinophil and glycoprotein responses were induced (Supplementary Figure 3 online). Thus, combined with histological confirmation of hyphal growth in the airways (Figure 3c, e and f), these observations confirm that conidia of A. niger produced an active airway infection that induced both allergic inflammation and airway obstruction even in the absence of demonstrable anti-fungal antibodies. Under these conditions we found no evidence of dissemination of A. niger to other organs (brain, liver, spleen; data not shown), hence infection was confined to the airway mucosal surface.
Fungal-dependent Th2 immunity against a bystander antigen enhances expression of allergic lung disease
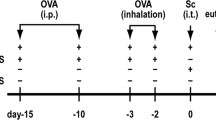
Concomitant with fungal conidia, which heavily contaminate homes (Table 1), humans are likely to inhale other environmental agents including innocuous antigens similar to ovalbumin that normally elicit only tolerogenic T-regulatory responses.15 We hypothesized that the allergic inflammation induced by low-level A. niger conidia exposure would be sufficient to abrogate T-regulatory responses and induce Th2-cell priming to this normally harmless antigen. To test this, groups of mice were challenged daily for 1 week with 5–10 × 103 A. niger conidia alone to establish a Th2-cell-predominant allergic immune lung environment, albeit one insufficient to elicit obstructive disease (Figure 5). Over the subsequent 2 weeks the mice were challenged simultaneously with conidia and ovalbumin.
Low-level exposure to viable conidia (5–10 × 103 per day) alone produced no significant airway hyperresponsiveness and only a modest degree of allergic inflammation despite substantial induction of airway glycoproteins and lung IL-4 secreting cells (Figure 6a–d). However, the addition of ovalbumin to ongoing low-level conidia challenge resulted in strong induction of AHR and a complete asthma phenotype, equivalent to exposure to 400 × 103 conidia alone, without markedly affecting other inflammatory indices (Figure 6a–d). Serum IgG1 reactivity against A. niger was not observed with these relatively low fungal infectious burdens and at least 20% of mice that received ovalbumin and low-grade fungal challenge had undetectable ovalbumin-specific IgG1 responses (Figure 6e).
A bystander antigen enhances A. niger-induced allergic lung disease. Mice received intranasally 5 × 103 (5 k) or 10 k A. niger conidia daily for 7 days after which mice continued to receive daily conidia with or without ovalbumin (OVA; 25 μg day–1) for an additional 14 days. Airway hyperresponsiveness (a), total BAL fluid-secreted glycoproteins (b) total BAL fluid (BALF) inflammatory cells (c), and total lung IL-4 and IFN-γ-secreting cells (d) were quantitated as in Figure 2. (e) The percent of the same mice with IgG1 reactivity to A. niger antigen or ovalbumin as assessed by western blot. Data are from one of two comparable experiments, n=3 mice/group. Panel a *P<0.05 5 k vs. OVA; **P<0.05 10 k vs. OVA Panels b–e *P<0.05 relative to OVA. BAL, bronchoalveolar lavage.
To determine whether ovalbumin-specific Th2-cell responses were generated by the addition of ovalbumin to low-level conidia challenge, mediastinal lymph node- and spleen-derived lymphocytes were restimulated with whole ovalbumin and the major histocompatibility complex class II-restricted ovalbumin323−339 peptide specific for mouse T-helper cells (Figure 7). Mice that had received only conidia failed to respond to either whole ovalbumin or ovalbumin peptide; moreover, mice receiving only intranasal ovalbumin failed to respond to ovalbumin restimulation in vitro (Figure 7). In contrast, in a dose-dependent manner, all mice receiving conidia and ovalbumin showed progressively more robust ovalbumin-specific IL-4 and IFN-γ responses in both lymph nodes and spleens. Peptide-stimulated cells confirmed that a predominant ovalbumin-specific Th2-cell response was generated under these conditions. Thus, low-grade allergic lung inflammation induced by A. niger conidia also induced robust Th2-cell activation against a normally tolerogenic bystander antigen even when antibody responses to the same antigen were not detectable. Moreover, such reactivity was physiologically relevant as the bystander Th2 response to ovalbumin was essential for full disease expression.
A. niger airway infection induces a Th2 response against a normally innocuous antigen. Leukocyte suspensions prepared from mediastinal lymph nodes (MLN; a,b) and spleens (c,d) removed from mice treated as in Figure 7 were restimulated in vitro with media alone, OVA323−339 peptide or whole ovalbumin (OVA) and the number of (a,c) IL-4- and (b,d) IFN-γ-secreting cells were determined by ELISpot. MLN data represent a single experiment involving pooled cells from five mice. *P<0.05 5 k vs. 5 k+OVA; **P<0.05 10 k vs. 10 k+OVA within same treatment group.
Fungal proteinases are required for fungal infection-dependent allergic lung disease
The preceding studies showed that contained low-grade fungal airway infection can induce allergic lung disease in a manner similar to fungal proteinases alone.14 To determine if allergenic fungi require secreted proteinases to induce allergic lung disease, we infected mice with wild-type A. niger (AB4.1) and two mutant A. niger strains deficient in either aspergillopepsin I (AB1.1) or multiple secreted proteinases (AB1.13), hyphae of the latter of which produce no detectable proteinase activity (data not shown),28 and determined the allergic lung disease phenotype (Figure 8). Airway hyperresponsiveness was only observed in mice infected with wild-type A. niger, although BAL fluid glycoprotein content was similar from all three mouse groups (Figure 8a and b). Furthermore, both the degree of airway eosinophilia and recruitment to lung of IL-4- and IFN-γ-secreting cells were strongly dependent on the amount of proteinase produced by the infecting fungal strain (Figure 8c and d). Indeed, the ratio of IL-4 to IFN-γ-secreting cells was profoundly affected by the degree to which infecting fungi produced proteinase (Figure 8d, inset). Clearance of fungal strain AB1.13 (no secreted proteinase) from lungs was not different from the wild-type fungal strain, but clearance of strain AB1.1 (aspergillopepsin-deficient) was markedly impaired at 24 h (Figure 8e). Moreover, after 2 weeks of conidia challenge, similar fungal colony forming units (CFU) were recovered from lung homogenates of mice challenged with wild-type (AB4.1) and proteinase null (AB1.13) strains, but approximately 80% greater CFU were recovered from mice challenged with strain AB1.1 (data not shown). These studies confirm that a complete asthma phenotype induced by fungal airway infection requires the secretion of proteinase by the fungus and rule out a requirement of the same proteinases for fungal growth in vivo. Secretion of aspergillopepsin was further required for optimal fungal clearance in vivo.
Proteinase production is required for A. niger-dependent allergic lung disease and fungal clearance. Mice received intranasally 400 × 103 A. niger conidia derived from wild-type (AB4.1), aspergillopepsin I-deficient (AB1.1) or multiple proteinase-deficient (AB1.13) strains every other day for a total of eight challenges. Data were collected 24 h after the final challenge. Airway hyperresponsiveness (a), total secreted BAL fluid (BALF) glycoproteins (b), total BAL fluid inflammatory cells (c) and total lung IL-4 and IFN-γ-secreting cells as assessed from whole lung (d; inset: IL-4/IFN-γ ratio) were determined as in Figure 2. Data are from one of two comparable experiments with n=5 animals each. Percentages refer to the percentage of proteinase activity present in hyphae relative to wild type (AB4.1) *P<0.05 comparing AB4.1–AB1.13; **P<0.05 comparing AB4.1–AB1.1. (e) Mice received 400 × 103 A. niger conidia intranasally once and the kinetics of fungal clearance from whole lung over 48 h were determined. Data are expressed as the percentage of CFU remaining relative to the initial 30-min time point. *P<0.05 relative to AB4.1 (wild-type strain). BAL, bronchoalveolar lavage; CFU, colony forming units.
Multiple components of the allergic inflammatory response are required for fungal clearance
If asthma-like disease is a common response to airway infection with proteinase-producing fungi, we reasoned that rather than being maladaptive and aberrant as is widely assumed,29 allergic lung disease might be protective against potentially lethal invasive fungal airway disease. To establish this, experiments were performed to assess the clearance of A. niger conidia from mice with pre-existing allergic lung disease and to determine the requirement of the key allergic effector moieties IL-5, eosinophils and IL-13, for fungal clearance (Figure 9). Mice with pre-existing allergic lung disease cleared A. niger conidia significantly faster relative to naive animals (Figure 9a). More rapid fungal clearance in this setting cannot be attributed to specific immune recognition of the fungus as the allergic lung disease was induced by an A. oryzae proteinase.14 Mice homozygous null for the IL-13 gene were impaired in their ability to clear A. niger infection (Figure 9b). Similarly, relative to control IgG-treated animals, wild-type mice administered a neutralizing anti-IL-5 antibody, which reduced airway eosinophilic responses to conidia challenge >95% (data not shown), were markedly impaired in their ability to clear A. niger infection (Figure 9c).
Allergic inflammation protects against fungal airway infection. (a) Mice were administered a fungal proteinase (A. oryzae) intranasally to elicit allergic lung disease or kept naive after which both groups received one intranasal challenge with 400 × 103 A. niger (AN) conidia. Lungs were removed at the indicated time points and fungal colony forming units (CFU) were determined and expressed as a percentage of naive control at the 0 time point. N=3 mice. (b) Naive wild-type and IL-13 null (–/–) mice received 400 × 103 A. niger conidia intranasally and clearance of the organism from whole lung was determined over 48 h. (c) Wild-type mice received once interperitoneally 0.5 mg of either an irrelevant control rat IgG antibody or a neutralizing anti-IL-5 antibody (TRFK5) and 24 h later received A. niger conidia intranasally as in b. Fungal clearance was then determined from whole lung over 48 h. N=5 mice. (d–f), 4 × 103 A. niger conidia were incubated in vitro with unfractionated BAL fluid cells (d) or highly purified eosinophils (e) obtained from proteinase-challenged mice at the indicated ratios and CFU were determined after 24 h and expressed as a percentage of the total CFU recovered in the absence of added BAL fluid cells. Data are from three experiments with four wells/condition. (f) Representative photomicrographs (× 400) from (e) illustrating fungal growth (black filaments) at different eosinophil/conidia ratios after 48 h of culture. *P⩽0.05.
The latter experiment suggested that eosinophils possessed anti-fungal properties. To prove this, eosinophil-enriched bronchoalveolar lavage fluid cells or highly purified bronchoalveolar lavage fluid eosinophils were incubated in vitro with viable A. niger conidia and the effect on fungal growth was determined (Figure 9d–f). Unfractionated BAL fluid cells showed substantial anti-fungal activity that was dependent on the ratio of BAL fluid cells to conidia (Figure 9d). However, highly purified eosinophils (>99%) were much more potent in this regard (Figure 9e; ratio of eosinophils/conidia required for a 90% suppressive effect 200:1 vs. 40:1, respectively; Figure 9d and e). At an eosinophil/ conidia ratio of 100:1, eosinophils were able to completely suppress all fungal growth (Figure 9f). Thus, multiple components of the allergic response to fungal airway infection contribute to the eradication of the infection and eosinophils specifically show potent anti-fungal activity.
Discussion
The discovery of allergenic proteinases provided the first insight into the molecular processes by which specific organisms elicit predominant Th2 responses and allergic lung inflammation linked to diseases such as asthma. Because fungi are strongly linked to asthma, ubiquitous in human environments and powerful producers of allergenic proteinases, they are compelling candidate initiators of asthma through their secreted proteinases. However, the precise relationship between fungi, proteinases, and asthma remains undefined. To clarify this, we initiated a detailed analysis of household dust to determine the existence and identity of active proteinases in relevant human environments. We have shown that common household fungi are a major source of active proteinases present in dust and that other known proteinases such as Der p 1 exist in enzymatically inactive form. Moreover, we showed that A. niger is very common, if not ubiquitous, in household dust and produced the most commonly observed proteinase. A. niger conidia readily infected the airways of C57BL/6 mice and elicited robust allergic inflammation and a disease syndrome strongly resembling allergic asthma often in the absence of detectable fungus-specific antibodies. Furthermore, we showed that far fewer conidia were required to induce allergic lung disease if a normally tolerogenic bystander antigen was administered concomitantly to the airways. Fungal-induced allergic lung disease required fungi capable of secreting proteinases and both IL-13 and eosinophils elicited during allergic lung disease were required for optimal eradication of fungal airway infection. These findings suggest that the relationship of fungi to asthma is based not simply on hypersensitivity to fungal products, but rather on active airway infection because of proteinase-producing household fungi and that asthma may be a protective response against fungal infection.
Because of the large number of allergenic organisms potentially present in household environments, we expected to detect by zymography, but not identify, many proteinases because of the lack of specific reagents. A major exception to this is Der p 1, which has been well characterized and for which specific detecting reagents are available. It was surprising, therefore, to find no Der p 1 proteinase activity despite detection of the protein in our dust samples (Table 1). This suggests that the catalytic activity of Der p 1 was unable to survive even in the relatively controlled environments of our study homes. Proteinase activity is required for full allergenicity, at least for fungal proteinases,14 therefore Der p 1 and other inactive household proteinases are less likely to participate as adjuvant-like initiators of allergic disease, although they could serve as bystander antigens that exacerbate allergic lung inflammation much like ovalbumin as used in our infectious model. This interpretation is consistent with previous epidemiological observations indicating that Der p 1 may act as a secondary, but not primary cause of asthma.30 Thus, rather than Der p 1, our findings indicate that active fungal proteinases produced during airway fungal infection may be the principal disease-initiating factor for many patients.
The most commonly isolated proteinase from our dust samples was approximately 86 kDa in size, which we repeatedly linked to A. niger. Although our detecting method is not definitive, it is likely that this dust proteinase is aspergillopepsin I (aspergillopeptidase A; extracellular acid protease; proteinase B), which has been variously reported to exist as ∼22 and ∼43 kDa proteins.28, 31, 32 Microbial proteinases are known to multimerize in solution and it is likely that our ∼86 kDa product represents a tetramer of aspergillopepsin I33 (Figure 1; and P Kolattukudy, personal communication). Aspergillopepsins are secreted by many aspergillus species and we have used the highly allergenic A. oryzae aspergillopepsin for many years to generate allergic lung disease in mice.14, 32
Both irradiated A. niger conidia and proteinase-deficient A. niger mutant conidia produced similar incomplete phenotypes in which mice failed to develop airway hyperreactivity, showed less airway eosinophilia and did not manifest the highly polarized Th2 response (lung IL-4/IFN-γ ratio >40:1) that are all characteristic of the complete disease phenotype seen after infection with wild-type fungi (Figures 2 and 8). Irradiated conidia are similar to proteinase-mutant A. niger as they represent an abundant source of proteinase-deficient chitin that has been shown to elicit innate IL-4 responses from mice.34 However, although irradiated conidia failed to germinate, conidia from proteinase-mutant A. niger strains showed robust growth, exceeding even that of the corresponding wild-type fungus (Figure 8). Therefore, proteinases were required for a complete disease phenotype induced by either conidia or mature organisms, but were not required for fungal growth either in vitro or in vivo. Together, these findings confirm that fungal proteinases are crucial virulence factors for allergic lung disease induced by fungal airway infection. The relatively low IL-4/elevated IFN-γ lung response induced by irradiated conidia and proteinase-deficient A. niger is a cytokine profile more consistent with the distinct lung syndrome of hypersensitivity pneumonitis and not asthma.35 Therefore, a potentially critical factor that distinguishes these distinct antigen-driven lung syndromes may be the degree of associated proteinase exposure or concomitant infection with fungi.
Although asthma is viewed as an aberrant immune reaction, our studies indicate that this perception is incorrect. Rather, asthma-like reactions most likely evolved to provide protection against host invasion by filamentous fungi (Figure 9). This establishes a duality for allergic lung inflammation, which may now be viewed as both harmful and potentially beneficial. This concept is perhaps most strikingly exemplified by the Th2 cytokine IL-13, which directly mediates airway obstruction during allergic lung disease,9 and yet as shown here provides substantial protection by enhancing fungal clearance (Figure 9). Numerous genetic polymorphisms have been linked to allergic asthma, all of which have been interpreted in the context of the harmful consequences of asthma. Our findings mandate that genetic studies of asthma be broadened to consider the potential beneficial effect of target genes on the airway response to environmental proteinases and fungi.
The precise relationship of atopy, the predilection to developing hypersensitivity reactions to environmental antigens, and asthma is not fully understood, but atopy is thought to be a primary, genetically controlled immune state that may lead to secondary disorders such as asthma.36, 37 Atopy is typically assessed by antibody-dependent reactions to specific allergens. However, our data show that mice often failed to generate detectable antibody responses to both fungi and bystander allergens such as ovalbumin (Figures 5 and 6), whereas T-cell responses to bystander antigens were fully preserved (Figure 7). This is consistent with our previous observations that allergic lung disease induced by fungal allergens is dependent on Th2 cells and not antibodies.38 Moreover, these findings indicate that antibody-based methods for detecting low-grade airway fungal infections and significant responses to bystander antigens (that is, atopy) lack sensitivity, at least in mice. This suggests that, using standard criteria, symptomatic human respiratory illnesses such as allergic sinusitis and asthma may be immunologically silent even if caused by an underlying fungal infection.39, 40 Clearly, a challenge of future studies designed to test novel associations between common fungi and human allergic illnesses will be to develop more sensitive diagnostic approaches.
Our study further suggests that atopy is not necessarily a primary disturbance, but may be a secondary response to low-grade airway fungal infection. Because of the ubiquitous nature of fungi, with a striking preponderance of Aspergillus and Penicillium spp. in human environments,41, 42 airway fungal infections are probably inescapable for humans and most other mammals. However, atopy to fungi is not common, either in the general population or in asthma subjects specifically. Nonetheless, subjects who show atopy to both fungi and other antigens have an increased incidence of respiratory tract allergic disease (48%).43 The hierarchical relationship between ubiquitous environmental fungi, fungal atopy, and symptomatic allergic disease precisely parallels our observations with A. niger-infected mice, further suggesting that such associations in human subjects are based on actual infection with proteinase-producing fungi and not merely hypersensitivity to fungal products. Because this represents a profoundly different concept of allergic disease pathogenesis, our studies further warrant definitive studies that dissect the multiple possible associations between environmental fungi and human diseases such as asthma.
Methods
Mice. C57BL/6J mice were purchased from Jackson Laboratories, Bar Harbor, ME. IL-13 null (–/–) mice were obtained from DNAX Research Institute, bred eight generations onto the C57BL/6 background and matched to wild-type littermate control animals. All mice were bred and housed at the American Association for Accreditation of Laboratory Animal Care-accredited vivarium at Baylor College of Medicine under specific pathogen-free conditions. All experimental protocols were approved by the Institutional Animal Care and Use Committee of Baylor College of Medicine and followed federal guidelines. Human studies were conducted according to Institutional Review Board protocols established at Baylor College of Medicine and the University of Texas Health Science Center at Houston and followed all federal guidelines.
House dust collection, aqueous extraction and analysis. Household dust was collected by a technician from asthmatic children's homes as part of the National Urban Air Toxics Research Center-funded study EPA R828678-01/CDFA 66.500.44 A total of five separate dust samples were collected from each house including the floor under subjects’ beds. A dust sleeve was placed into the hose of a standard portable vacuum cleaner to collect samples. Vacuuming was performed for 4 min and confined to a 4 ft2 area defined by an open template placed in appropriate locations. Dust sleeves were then sealed in plastic bags, packed on ice and stored at −20 °C within 24 h to preserve proteolytic and biological activity. A maximum of 84 dust samples obtained from 31 homes were available for this study from which the following determinations were made: Der p 1 concentration, conidia count, endotoxin analysis, total proteinase activity, and zymogram. Approximately 0.5–2 g of dust were wetted with ∼3 ml g–1 phosphate-buffered saline (PBS). The suspension was passed though a 45 μm mesh (BD Falcon; Bedford, MA) to remove large debris. Samples were centrifuged at 10,000 × g for 5 min and the supernatant was removed and stored at −80 °C From these aqueous samples, total proteolytic activity was determined by EnzChek (Invitrogen/Molecular Probes; Carlsbad, CA) as described.14 Total conidia counts, total endotoxin, and Der p 1 concentrations were determined by the Johns Hopkins University Dermatology, Allergy and Clinical Immunology (DACI) Reference Laboratory (http://www.hopkinsmedicine.org/allergy/daci/index.html). Limiting dust quantities precluded making all determinations for all samples (Table 1). Proteinase activity of purified Der p1 (Indoor Biotechnologies, Charlottesville, VA) and aqueous house dust extracts was retrieved by incubating in phosphate-buffered saline (PBS; pH 7.5) containing 1 mM EDTA and 1 mM DTT for a minimun of 1 h at 37 °C. Zymography was then performed using 10% SDS-polyacrylamide gel Electrophoresis (SDS-PAGE) as described (Kheradmand, 2002 no. 4576).
Fungi and fungal isolation. The insoluble dust material remaining following PBS extraction was resuspended in ∼2 ml PBS per gram dust. 2, 10, and 100 μl aliquots were plated on Sabouraud's media (Becton Dickinson, Sparks, MD) supplemented with 50 μg ml–1 ampicillin and incubated at 37 °C for up to 5 days. A. niger and other species were identified according to typical appearance in culture and by microscopic characteristics with results confirmed by Microcheck (Northfield, VT).
Mature, heavily sporulated fungal isolate plate cultures were washed repeatedly with PBS 1% Tween 80 after which conidia suspensions were passed through 45 μm nylon mesh to remove hyphae. Suspensions were washed twice with PBS by centrifugation (10,000 × g, 5 min, 4 °C) and the pellets were resuspended in PBS. Conidia concentration was determined using a hemacytometer and adjusted to 8.0 × 107 conidia per ml and aliquots were stored in liquid nitrogen to preserve conidia viability. Conidia viability after freezing was confirmed by determining colony forming ability (CFA) by plating serial dilutions on Sabouraud's media. In vivo experiments were performed with conidia from a single A. niger isolate obtained from house dust (available from authors). Proteinase-mutant A. niger strains (AB1.1 and AB1.13) and the parental wild-type strain (AB4.1) were obtained from TNO (Delft, The Netherlands; http://www.TNO.nl) and grown as directed. AB1.1 contains a site directed deletion of aspergillopepsin I and AB1.13 is a UV light-induced mutant with no measurable secreted proteinase activity.28
Irradiation of fungal conidia. Conidia collected from plates as above were centrifuged (10,000 × g for 5 min, 4 °C) and PBS supernatant was aspirated. The pellet was exposed to 3 Gy of γ-radiation (0.5 Gy per hour) on ice. Conidia were resuspended in PBS at 8 × 107 conidia per ml. Irradiated conidia contained <1 viable conidium per 107; non-irradiated preparations contained ∼35 viable conidia per 102.
Antigens and intranasal challenge. Chicken egg ovalbumin (OVA; Sigma Chemical, St Louis, MO) was reconstituted in sterile PBS at 500 μg ml–1 and passed serially through polymyxin B columns until endotoxin content was <0.01 EU μg–1 and stored at −80 °C. The A. oryzae aspergillopepsin was combined with ovalbumin and administered to mice to elicit allergic lung disease as previously described.14 For A. niger-derived antigen, Sabourad's broth supplemented with 50 μg ml–1 ampicillin was inoculated with 4 × 106 conidia per 100 ml and cultures kept in a shaking incubator for 48 h at 37 °C to prevent conidia production. Hyphae were collected, washed twice with PBS, placed on ice and ground in a PM100 planetary ball mill (Retsch; Newtown, PA) using 2 mm balls. The resulting colloid was centrifuged at 10,000 × g for 30 min at 4 °C to remove particulate material, the supernatant protein concentration was determined by BCA method (Thermo Fisher Scientific, Rockford, IL) and aliquots were stored at −20 °C. For intranasal challenge, aliquots of conidia were diluted in PBS to achieve the indicated number of conidia per 50 μl volume. Mice were deeply anesthetized with isoflurane and droplets containing PBS, conidia in PBS or ovalbumin were applied to the nares until a total of 50 μl was inhaled. Similar studies using this protocol with Streptococcus pneumoniae indicated that 87–95% of the colony forming units administered to the nares were immediately recoverable from the lungs (D Corry, J Prince, data not shown).
Quantitation of allergic lung disease. Bronchial responsiveness to acetylcholine (Ach) intravenous challenge, BAL fluid collection, quantitation of airway inflammatory cells, and secreted glycoproteins, analysis of total lung IL-4 and IFN-γ-secreting cells, and lung histopathology were carried out as previously described.14, 38, 45 Quantitation of cytokines from BAL fluid was performed by bead-assisted analysis (MILLIPLEX MAP Kit Mouse Cytokine/Chemokine Immunoassay; Millipore, Billerica, MA, USA) using a Bioplex analyzer (BioRad, Hercules, CA) according to the manufacturers’ protocols.
Western blotting. Standard SDS-PAGE was performed using 100 μg of fungal antigen or 5 μg of chicken egg OVA. After transfer to nylon membranes, even protein transfer was confirmed using Ponceau red (Sigma). While stained, membranes were cut into separate strips in order to test individual serum samples. Strips were blocked 1 h in TBST (10 mM Tris-HCl pH 7.4, 150 mM NaCl, 0.1% Tween 20) containing 5% (w/v) non-fat dehydrated milk (Carnation non-fat dry milk (Nestlé, Solon, OH)). Strips were incubated 1 h at room temperature with a 1:200 dilution of mouse serum in TBST with 5% milk. The membrane strips were washed with TBST and incubated at room temperature with a 1:10,000 dilution of rat anti-mouse IgG1 or IgE antibody conjugated to HRP (BD Pharmingen, San Jose, CA) in TBST with 5% milk. Strips were washed in TBST repeatedly and signal was detected using enhanced chemiluminescence substrate (Pierce, Rockford, IL).
Conidia clearance assay. Mice received a single intranasal challenge with 400 × 103 A. niger conidia after which BAL fluid and lungs were collected at 30 min, 1, 2, 3, 5, 7 and 14 days after challenge. Single-cell suspensions of lungs were prepared by pressing through 45 μm mesh and aliquots of BAL fluid and lung-cell suspensions were plated on Sabourad's ampicillin plates and placed at 37 °C CFU were enumerated 24 h after plating and cultures were allowed to mature to sporulation to confirm identity of A. niger. Clearance data are presented as combined BAL and lung homogenate CFU and reported as the percent remaining relative to the 30 min time point. For some clearance studies, mice were administered once intraperitoneally 0.5 mg of a neutralizing anti-IL-5 antibody (TRFK5 (Schumacher, 1988 no. 427)) 24 h before conidia challenge. This antibody dose was sufficient to reduce lung eosinophilia in response to conidia >95% (data not shown).
Antigen-specific leukocyte restimulation. Spleens and mediastinal lymph nodes were collected from conidia- and ovalbumin-challenged animals 1 week following cessation of all intranasal challenges. Organs were lightly pressed through 45 μm nylon mesh to create single-cell suspensions, which were washed once in complete medium (RPMI 1640 medium supplemented with 10% heat inactivated fetal calf serum, penicillin, streptomycin, and glutamine). Cell suspensions were added to flat bottom wells of high-protein-binding microtiter plates (Immulon II; Dynatech, Chantilly, IL) that had been pre-coated with anti-IL-4 (11B11; BD Pharmingen, San Diego, CA) or anti-IFN-γ (AN18; gift of Dr Anne O’Garra) antibodies. PBS, ovalbumin (1 mg ml–1) or OVA323−339 peptide (ISQAVHAAHAEINEAGR; 10 μg ml–1) were then added and cultures were continued for 24 h, after which plates were developed per ELISpot protocol and the total number of IL-4 and IFN-γ-secreting cells were determined as described.14
Fungal killing assay. Mice were challenged intranasally with 7 μg A. oryzae proteinase (Sigma) and 22 μg ovalbumin every other day for 16 days to elicit ∼90% BAL fluid eosinophilia. BAL fluid was collected as above to harvest airway cells, which were incubated in increasing numbers with 4 × 103 A. niger conidia in complete medium (RPMI 1640, 2 mM L-glutamine, 100 units ml–1 penicillin, 100 μg ml–1 streptomycin, and 10% fetal bovine serum (Hyclone Logan, Utah)). Cultures were incubated for 24 h at 37 °C in a room air cell culture incubator after which fungal colonies were counted directly from culture plates. Cultures were incubated for an additional 24 h for documentation of hyphal growth. Additional experiments were performed otherwise identically using >99% pure eosinophils isolated by flow cytometry as previously described.46
Statistical analysis. Data are presented as means±s.e.m.. Significant differences are expressed relative to control mice (P⩽0.05) using Student's t-test assuming two-tailed distribution and unequal variance.
References
Grant, E.N., Wagner, R. & Weiss, K.B. Observations on emerging patterns of asthma in our society. J. Allergy Clin. Immunol. 104, S1–S9 (1999).
Mannino, D.M. et al. Surveillance for asthma-United States, 1980–1999. MMWR Surveil Summ. 51, 1–13 (2002).
Gratziou, C. et al. Inflammatory and T-cell profile of asthmatic airways 6 h after local allergen provocation. Am. J. Respir. Crit. Care. Med. 153, 515–520 (1996).
Humbert, M. et al. IL-4 and IL-5 mRNA and protein in bronchial biopsies from patients with atopic and nonatopic asthma: evidence against “intrinsic” asthma being a distinct immunopathologic entity. Am. J. Respir. Crit. Care. Med. 154, 1497–1504 (1996).
Kraft, M., Djukanovic, R., Wilson, S., Holgate, S.T. & Martin, R.J. Alveolar tissue inflammation in asthma. Am. J. Respir. Crit. Care. Med. 154, 1505–1510 (1996).
Robinson, D.S. et al. Predominant TH2-like bronchoalveolar T-lymphocyte population in atopic asthma. N. Engl. J. Med. 326, 298–304 (1992).
Synek, M. et al. Cellular infiltration of the airways in asthma of varying severity. Am. J. Respir. Crit. Care. Med. 154, 224–230 (1996).
Molfino, N.A., Nannini, L.J., Martelli, A.N. & Slutsky, A.S. Respiratory arrest in near-fatal asthma. N. Engl. J. Med. 324, 285–288 (1991).
Grunig, G. et al. Requirement for IL-13 independently of IL-4 in experimental asthma. Science 282, 2261–2263 (1998).
Corry, D.B. et al. Interleukin 4, but not interleukin 5 or eosinophils, is required in a murine model of acute airway hyperreactivity. J. Exp. Med. 183, 109–117 (1996).
Busse, W.W., Coffman, R.L., Gelfand, E.W., Kay, A.B. & Rosenwasser, L.J. Mechanisms of persistent airway inflammation in asthma. A role for T cells and T-cell products. Am. J. Respir. Crit. Care. Med. 152, 388–393 (1995).
Krinzman, S.J. et al. T cell activation in a murine model of asthma. Am. J. Physiol. 271, L476–83 (1996).
Angkasekwinai, P. et al. Interleukin 25 promotes the initiation of proallergic type 2 responses. J. Exp. Med. 204, 1509–1517 (2007).
Kheradmand, F. et al. A protease-activated pathway underlying Th cell type 2 activation and allergic lung disease. J. Immunol. 169, 5904–5911 (2002).
Akbari, O. et al. Antigen-specific regulatory T cells develop via the ICOS-ICOS-ligand pathway and inhibit allergen-induced airway hyperreactivity. Nat.Med. 8, 1024–1032 (2002).
Heaton, T. et al. An immunoepidemiological approach to asthma: identification of in-vitro T-cell response patterns associated with different wheezing phenotypes in children. Lancet 365, 142–149 (2005).
Dold, S., Heinrich, J., Wichmann, H.E. & Wjst, M. Ascaris-specific IgE and allergic sensitization in a cohort of school children in the former East Germany. J. Allergy Clin. Immunol. 102, 414–420 (1998).
Dormann, D. et al. Heterogeneity in the polyclonal T cell response to birch pollen allergens. Int. Arch. Allergy Immunol. 114, 272–277 (1997).
Sokol, C.L., Barton, G.M., Farr, A.G. & Medzhitov, R. A mechanism for the initiation of allergen-induced T helper type 2 responses. Nat. Immunol. 9, 310–318 (2008).
Lamhamedi-Cherradi, S.-E. et al. Fungal proteases induce Th2 polarization through limited dendritic cell maturation and reduced production of IL-12. J. Immunol. 180, 6000–6009 (2008).
Kiss, A. et al. A new mechanism regulating the initiation of allergic airway inflammation. J. Allergy Clin. Immunol. 120, 334–342 (2007).
Johnson, T.F., Reisman, R.E. & Arbesman, C.E. Late onset asthma due to inhalation of Aspergillus niger. Clin. Allergy 5, 397–401 (1975).
Hogaboam, C.M. et al. Chronic airway hyperreactivity, goblet cell hyperplasia, and peribronchial fibrosis during allergic airway disease induced by Aspergillus fumigatus. Am. J. Pathol. 156, 723–732 (2000).
Denis, O. et al. Chronic intranasal administration of mould spores or extracts to unsensitized mice leads to lung allergic inflammation, hyper-reactivity and remodelling. Immunology 122, 268–278 (2007).
Ward, G.W. Jr ., Karlsson, G., Rose, G. & Platts-Mills, T.A. Trichophyton asthma: sensitisation of bronchi and upper airways to dermatophyte antigen. Lancet 1, 859–862 (1989).
Ward, G.W. Jr ., Woodfolk, J.A., Hayden, M.L., Jackson, S. & Platts-Mills, T.A. Treatment of late-onset asthma with fluconazole. J. Allergy Clin. Immunol. 104, 541–546 (1999).
Ward, R.E. & McLaren, D.J. Schistosoma mansoni: migration and attrition of challenge parasites in naive rats and rats protected with vaccine serum. Parasite Immunol. 11, 125–46 (1989).
Mattern, I.E. et al. Isolation and characterization of mutants of Aspergillus niger deficient in extracellular proteases. Mol. Gen. Genet. 234, 332–336 (1992).
Wills-Karp, M. Immunologic basis of antigen-induced airway hyperresponsiveness. Annu. Rev. Immunol. 17, 255–281 (1999).
Pearce, N., Douwes, J. & Beasley, R. Is allergen exposure the major primary cause of asthma? Thorax 55, 424–431 (2000).
Takahashi, K. et al. The primary structure of Aspergillus niger acid proteinase A. J. Biol. Chem. 266, 19480–19483 (1991).
Ichishima, E. 294. Aspergillopepsin I. In Handbook of Proteolytic Enzymes (Barrett, A.J., Rawlings, N.D. & Woessner, J.F. eds) 872–878 ( Academic Press, San Diego, 1998 ).
Pacaud, M., Sibilli, S. & Bras, G. Protease I from Escherichia coli. Some physicochemical properties and substrate specificity. Eur. J. Biochem. 69, 141–151 (1976).
Reese, T.A. et al. Chitin induces accumulation in tissue of innate immune cells associated with allergy. Nature 447, 92–96 (2007).
Gudmundsson, G. & Hunninghake, G.W. Interferon-gamma is necessary for the expression of hypersensitivity pneumonitis. J. Clin. Invest. 99, 2386–2390 (1997).
Postma, D.S. et al. Genetic susceptibility to asthma--bronchial hyperresponsiveness coinherited with a major gene for atopy. N. Engl. J. Med. 333, 894–900 (1995).
Daniels, S.E. et al. A genome-wide search for quantitative trait loci underlying asthma. Nature 383, 247–50 (1996).
Corry, D.B. et al. Requirements for allergen-induced airway hyperreactivity in T and B cell-deficient mice. Mol. Med. 4, 344–355 (1998).
Black, P.N., Udy, A.A. & Brodie, S.M. Sensitivity to fungal allergens is a risk factor for life-threatening asthma. Allergy 55, 501–504 (2000).
Ezeamuzie, C.I. et al. IgE-mediated sensitization to mould allergens among patients with allergic respiratory diseases in a desert environment. Int. Arch. Allergy Immunol. 121, 300–307 (2000).
Ceylan, E. et al. Fungi and indoor conditions in asthma patients. J Asthma 43, 789–94 (2006).
Cetinkaya, Z. et al. Assessment of indoor air fungi in Western-Anatolia, Turkey. Asian Pac. J. Allergy Immunol. 23, 87–92 (2005).
Katz, Y. et al. Indoor survey of moulds and prevalence of mould atopy in Israel. Clin. Exp. Allergy 29, 186–192 (1999).
Delclos, G.L. et al. Oxygenated urban air toxics and asthma variability in middle-school children: A panel study. in NUATRC Res Rep ( Mickey Leland National Urban Air Toxics Research Center, Houston, TX, 2007 ).
Xu, J., Park, P.W., Kheradmand, F. & Corry, D.B. Endogenous attenuation of allergic lung inflammation by syndecan-1. J. Immunol. 174, 5758–5765 (2005).
Montes, M., Jaensson, E.A., Orozco, A.F., Lewis, D.E. & Corry, D.B. A general method for bead-enhanced quantitation by flow cytometry. J. Immunol. Methods 317, 45–55 (2006).
Acknowledgements
Supported by NIH Grants HL75243 and AI057696 (to DBC), AI070973 (to GLD, SA, FK and DBC) and AI07495 (to PP).
Author information
Authors and Affiliations
Corresponding author
Additional information
DISCLOSURE
The authors declared no conflict of interest.
SUPPLEMENTARY MATERIAL is linked to the online version of the paper at http://www.nature.com/mi
Supplementary information
Rights and permissions
About this article
Cite this article
Porter, P., Susarla, S., Polikepahad, S. et al. Link between allergic asthma and airway mucosal infection suggested by proteinase-secreting household fungi. Mucosal Immunol 2, 504–517 (2009). https://doi.org/10.1038/mi.2009.102
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/mi.2009.102
This article is cited by
-
Novel acute hypersensitivity pneumonitis model induced by airway mycosis and high dose lipopolysaccharide
Respiratory Research (2021)
-
Studies of Atmospheric PM2.5 and its Inorganic Water Soluble Ions and Trace Elements around Southeast Asia: a Review
Asia-Pacific Journal of Atmospheric Sciences (2021)
-
Caspase-11 promotes allergic airway inflammation
Nature Communications (2020)
-
The External Exposome and Food Allergy
Current Allergy and Asthma Reports (2020)
-
Role of Environmental Adjuvants in Asthma Development
Current Allergy and Asthma Reports (2020)