Abstract
The mucosal tissues of the gastrointestinal, respiratory, reproductive, and urinary tracts, and the surface of the eye present an enormous surface area to the exterior environment. All of these tissues are covered with resident microbial flora, which vary considerably in composition and complexity. Mucosal tissues represent the site of infection or route of access for the majority of viruses, bacteria, yeast, protozoa, and multicellular parasites that cause human disease. Mucin glycoproteins are secreted in large quantities by mucosal epithelia, and cell surface mucins are a prominent feature of the apical glycocalyx of all mucosal epithelia. In this review, we highlight the central role played by mucins in accommodating the resident commensal flora and limiting infectious disease, interplay between underlying innate and adaptive immunity and mucins, and the strategies used by successful mucosal pathogens to subvert or avoid the mucin barrier, with a particular focus on bacteria.
Similar content being viewed by others
Mucins—an Integral Part of the Mucosal Barrier
Mucosal epithelial tissues have evolved multiple mechanisms of defense in response to their vulnerability to microbial attack due to their exposure to the external environment. The mucosal epithelial cells form a contiguous lining that acts as a barrier between the moist exterior environment and the remainder of the host. In addition, these cells, both constitutively and in response to microbes, together with underlying leukocytes, secrete many defensive compounds into the mucosal fluid, including mucins, antibodies, defensins, protegrins, collectins, cathlecidins, lysozyme, histatins, and nitric oxide.1, 2 and 3 Together, these different defensive compounds form a physical barrier and have direct antimicrobial activity, and the ability to opsonize microbes to aid clearance. Mucin glycoproteins, however, can fulfill all of these roles individually.
Mucosal pathogens, almost by definition, have evolved mecha-nisms to subvert these mucosal defensive measures. The first barrier the pathogen encounters is the highly hydrated mucus gel that covers the mucosal surface and protects the epithelial cells against chemical, enzymatic, microbial, and mechanical insult. Mucosal surfaces are coated with a layer of viscous mucus ranging in thickness from 10 μm in the eye4 and trachea5 to 300 μm in the stomach and 700 μm in the intestine.6, 7 and 8 This mucus layer is not static but moves to clear trapped material. In the gastrointestinal tract, the outer mucus layer is continually removed by movement of the luminal contents, whereas in the respiratory tract cilia drive its movement. Mucin glycoproteins produced by mucus-producing cells in the epithelium or submucosal glands are the major macromolecular constituent of mucus and are responsible for the viscous properties of the mucus gel. In addition to forming a relatively impervious gel, which acts as a lubricant, a physical barrier, and a trap for microbes, mucus provides a matrix for a rich array of antimicrobial molecules.
Underneath the mucus layer, the cells present a dense forest of highly diverse glycoproteins and glycolipids, which form the glycocalyx. Membrane-anchored cell-surface mucin glyco-proteins are a major constituent of the glycocalyx in all mucosal tissues. The glycocalyx is highly variable from tissue to tissue; for example, the glycocalyx of human intestinal microvilli tips is thick (100–500 nm) in comparison with the glycocalyx of the lateral microvilli surface (30–60 nm).9, 10 The oligosaccharide moieties of the molecules forming the glycocalyx and the mucus layer are highly diverse, and the average turnover time of the human jejunal glycocalyx is 6–12 h.11 Consequently, both the secreted and adherent mucosal barriers are constantly renewed and could potentially be rapidly adjusted to changes in the environment, for example, in response to microbial infection.
Mucin Biosynthesis and Structure
The tremendous energy investment by mucosal tissues in the production of mucins in the basal state, but particularly in response to infection, is testimony to the importance of these glycoproteins. Epithelial mucins are a heterogenous family of large complex glycoproteins containing a dense array of O-linked carbohydrates typically comprising over 70% of their mass. These glycans are concentrated in large peptide domains of repeating amino-acid sequences rich in serine and threonine. The size and number of repeats vary between the mucins, and in many of the genes there are genetic polymorphisms in the number of repeats (variable number of tandem repeats or VNTR polymorphisms), which means the size of individual mucins can differ substantially between individuals. Each mucin is thought to form a filamentous protein carrying typically 100s of complex oligosaccharide structures,12 giving the mucin a “bottle-brush” appearance. To date, at least 16 human mucins have been included in the family, and the expression profile of mucins varies between tissues with the gastrointestinal tract showing the highest and most diverse expression (see Table 1).
Mucins can be divided into three distinct subfamilies: (a) secreted gel-forming mucins, (b) cell-surface mucins, and (c) secreted non-gel-forming mucins (Table 1). Gel-forming mucins, which are the major constituent of mucus and confer its viscoelastic properties, are encoded by a cluster of four highly related genes on chromosome 1113, 14 and 15 and a similar gene on chromosome 12.16 Gel-forming mucins contain N- and C-terminal cysteine-rich domains that are both involved in homo-oligomerization mediated by inter-molecular disulfide bonds.17, 18 The current model for mucin oligomerization is that dimerization occurs rapidly during biosynthesis in the endoplasmic reticulum preceding or concomitant with N-glycosylation but before O-glycosylation in the Golgi apparatus, which in turn is followed by multimerization of dimers.19 Oligomerization is likely to produce either extended filamentous structures or, more probably, web-like molecular structures likely to be critical to the rheological properties of the mucus gel.20, 21, 22, 23 and 24 The extended conformation caused by dense glycosylation enables the molecules to occupy large volumes, with the secreted oligomeric mucins occupying volumes equivalent to those of small bacteria.25 The secreted non-oligomerizing mucins include the MUC7 salivary mucin, which can self-aggregate but is not thought to contribute significantly to mucus properties, and the MUC8 respiratory mucin, which has not been fully characterized to date.
There are 11 known genes encoding cell-surface mucins expressed by a wide diversity of mucosal tissues, with substantial redundancy in many tissues (see Table 1). Cell-surface mucins are present on the apical membrane of all mucosal epithelial cells and contain large extracellular VNTR domains predicted to form rigid elongated structures. Together with their high expression, this indicates that these molecules are likely to be a prominent, probably dominating, constituent of the glycocalyx and may provide a barrier that limits access of other cells and large molecules to the cell surface. During synthesis, most cell-surface mucins appear to be cleaved into two subunits in a region known as an SEA module, which is often flanked by epidermal growth factor-like domains.26 Structural studies of the MUC1 SEA module suggest that the cleavage occurs via autoproteolysis and that the two subunits remain non-covalently associated throughout biosynthesis.27 However, importantly, the extracellular α-subunit can be shed from the cell surface either mediated via a second distinct cleavage event28, 29 or perhaps via physical shear forces separating the two domains at the original cleavage site as suggested by Macao et al.27 Mutation of the cleavage site inhibits MUC1 shedding in transfected mammary and respiratory epithelial cells without affecting cell surface expression, indicating the importance of the initial cleavage in shedding.30 Furthermore, most cell-surface mucin genes appear to undergo alternative splicing and also encode directly secreted isoforms lacking transmembrane and cytoplasmic domains.31, 32, 33 and 34 These isoforms are stored in subapical granules and in goblet cell thecae and are secreted both constitutively and following stimuli. Consequently, due to shedding and direct secretion, cell-surface mucins can also be seen as components of secreted mucus.35
MUC1 is the most extensively studied membrane-associated mucin and is the most ubiquitously expressed across all mucosal tissues. MUC1 has been estimated to be 200–500 nm in length (depending on the number of tandem repeats), suggesting it will tower above other molecules attached to the plasma membrane.36 MUC1 associated with the cell surface is constantly internalized (0.9% of the surface fraction min−1) and recycled.37 Internalization occurs by clathrin-mediated endo-cytosis, and alterations in O-glycan structure stimulate endo-cytosis and intracellular accumulation.38 During recycling, sialic acid is added to the premature form of MUC1.37 Complete sialylation requires several rounds of recycling, one cycle taking approximately 2.5 h.37 Pulse-chase experiments indicate that the half-life of MUC1 in the plasma membrane is 16–24 h,37, 39 suggesting that the average MUC1 molecule recycles up to 10 times before release.37 Recycling rates vary between cell lines and possibly environmental conditions and have not been measured in non-transformed cells, making it very difficult to extrapolate to the real rate of recycling and cell-surface half-life in vivo. The cytoplasmic tail appears to interact with the cytoskeleton and secondary signaling molecules,40, 41 and 42 whereas the extracellular domains of MUC1 and other cell-surface mucins interact with extracellular matrix components and other cells.43, 44, 45, 46 and 47
The cytoplasmic domains of the cell surface mucins are complex, often contain known phosphorylation motifs, and are highly conserved across species, suggesting important intra-cellular functions. The best-studied mucin in this regard is MUC1, which has been explored mainly in terms of its role in cancer cell biology rather than in mucosal defense. We and others have shown phosphorylation of the MUC1 cytoplasmic domain41, 48, 49, 50, 51 and 52 as well as molecular association with β-catenin,41, 51 linking MUC1 with the Wnt pathway, which is involved in epithelial growth, migration, and wound repair. More recently, it has been shown that the cytoplasmic domain can be cleaved and that the cleaved domain translocates to mitochondria and, together with the p53 transcription factor, to the nucleus, where it modulates the cell cycle and protects against the apoptotic response to genotoxic stress.53, 54 Some pathogenic bacteria produce genotoxins, and thus this protective effect, first identified in cancer cells, may have evolved as part of the natural epithelial defense against microbial genotoxins. We have recently shown that in vitro MUC1 protects p53-expressing epithelial cells from the effects of cytolethal distending toxin, a genotoxin produced by Campylobacter jejuni.55, 56In vivo, C. jejuni more densely colonized the stomachs of Muc1−/− mice, but this effect was not seen in isogenic mutants lacking cytolethal distending toxin, indicating that Muc1 lowers gastric colonization at least in part via inhibiting the activity of cytolethal distending toxin.55 Many of the other cell-surface mucins also contain potential phosphorylation sites and cleavage motifs in the immediate intracellular region of their cytoplasmic domains and may be similarly cleaved. Importantly, there is also evidence that interaction with bacteria can induce phosphorylation of MUC1 in vitro.57 Signaling by the cytoplasmic domains of cell-surface mucins is complex and much remains to be elucidated about their mode of action. However, the evidence to date suggests that these domains are involved in cellular programs regulating growth and apoptosis in mucosal cells perhaps in response to microbes and/or their toxins.
Mucin Glycosylation
The carbohydrate structures present on mucosal surfaces vary according to cell lineage, tissue location, and developmental stage. Evidence is emerging that mucin glycosylation can alter in response to mucosal infection and inflammation, and this may be an important mechanism for unfavorably changing the niche occupied by mucosal pathogens. The extensive O-glycosylation of the mucins protects them from proteolytic enzymes and induces a relatively extended conformation.25 The oligosaccharides on secreted mucins are clustered into heavily glycosylated domains (typically 600–1,200 amino acids long) separated by shorter nonglycosylated regions.25 The O-linked glycans contain 1–20 residues, which occur both as linear and branched structures (see Table 2). The carbohydrate chain is initiated with an N-acetylgalactosamine (GalNAc) linked to serine or threonine and is elongated by the formation of the so-called core structures followed by the backbone region (type-1 and type-2 chains). The chains are typically terminated by fucose (Fuc), galactose (Gal), GalNAc, or sialic acid residues in the peripheral region, forming histo-blood-group antigens such as A, B, H, Lewis-a (Lea), Lewis-b (Leb), Lewis-x (Lex), Lewis-y (Ley), as well as sialyl-Lea and sialyl-Lex structures. Sulfation of Gal and N-acetylglucosamine (GlcNAc) residues causes further diversification. In addition to the O-linked glycans, mucins contain a smaller number of N-linked oligosaccharides, which have been implicated in folding, oligomerization (MUC2), or surface localization (MUC17).58, 59 and 60
The carbohydrate structures present on mucins are determined by the expression of specific glycosyl transferases. Thus, mucin glycosylation is governed by genetics (due to polymorphisms in these enzymes), tissue-specific enzyme expression, and host and environmental factors influencing transferase expression. As an example of the impact of host genotype, the H type-1 structure is made by the secretor (Se) gene product, and the majority (80% of Caucasians, all South American Indians and Orientals) carry this structure and are thus referred to as “secretors”.61 Individuals may also express the Lewis (Le) gene (90% of the Caucasian population) and, provided that they are also secretors, will modify H type-1 into the Leb structure.61, 62 If they are nonsecretors, type-1 chains without its blood group antigen H will be turned into Lea structures.61, 62 The third Se phenotype with simultaneous expression of Lea and Leb antigens has been described as the “weak-Secretor” (Sew) phenotype.63, 64 The dual expression of Lea/Leb is a consequence of an enzymatically weak Se-transferase in combination with an intact Le-transferase.63, 64 The terminal structures of mucin oligosaccharides are highly heterogeneous and vary between/within species and between and even within tissues. The array of oligosaccharide structures on individual mucin molecules is also somewhat determined by stochastic events as the mucin protein moves through the Golgi apparatus.65 This structural diversity may allow the mammalian host to cope with diverse and rapidly changing pathogens, as reflected by the observation that susceptibility to specific pathogens differs between people with different histo-blood groups,66 as exemplified by the observations that individual Se phenotype may determine the ratio of infection as well as the course and severity of urinary tract infections, Norwalk virus induced acute gastroenteritis and Helicobacter pylori-induced gastric diseases.67, 68 and 69 There is also a strong correlation between distinct adhesive properties of H. pylori endemic in specific human populations and the mucin blood group carbohydrate structures expressed by these populations.70 These differences in the external barrier to infection can be equated with the diversity in underlying innate and adaptive immunity (e.g., polymorphisms in MHC, cytokines), which is thought to have evolved for the same reasons.
Regulation and Modulation of the Mucin Barrier
The gel-forming mucins are produced by cells in the epithelial surface and/or by glands located in the submucosal connective tissues, and secretion occurs via both constitutive and regulated pathways.71 Both gel-forming and cell-surface mucins show constitutive and inducible gene expression in mucosal epithelial cells. The promoters of the MUC genes have generally not been fully characterized, although partial promoter characterization is available for human MUC1,72, 73 and 74MUC2,75, 76, 77, 78, 79, 80 and 81MUC3,82MUC4,83, 84 and 85MUC5AC,79, 86 and MUC5B.87 Differential regulation of individual mucin genes is evident between different mucosal tissues and throughout differing regions of the larger epithelial tracts. For example, differing promoter regions are involved in the differential regulation of constitutive MUC2 expression in the small and large intestines.78 Expression of cell-surface and gel-forming mucins can be upregulated by inflammatory cytokines such as interleukin (IL)-1β, IL-4, IL-6, IL-9, IL-13, interferons, tumor necrosis factor-α, nitric oxide, and other uncharacterized inflammatory factors.82, 88, 89, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99, 100, 101, 102, 103, 104, 105, 106, 107, 108, 109 and 110 Responsiveness to these cytokines provides a link between mucins, innate mucosal immunity, and mucosal inflammatory responses. Neutrophils can also stimulate increases in production of both gel-forming and cell-surface mucins by mucosal epithelial cells via neutrophil elastase.111, 112, 113, 114, 115 and 116 Microbial products can stimulate increased production of mucins by mucosal epithelial cells.103, 117, 118, 119 and 120 In fact, there is evidence that adherence of probiotic bacteria upregulates cell-surface mucin expression in vitro,121, 122 perhaps representing an important part of the mechanism by which probiotic bacteria limit infection by pathogens. In contrast, the lipopolysaccharide of the pathogen H. pylori decreases mucin synthesis in gastric epithelial cells in vitro via activation of cPLA-2,123 representing a mechanism by which a pathogen can favorably modulate the mucus barrier.
The constitutive pathway continuously secretes sufficient mucin to maintain the mucus layer, whereas the regulated pathway affords a massive discharge as a response to environmental and/or (patho)physiological stimuli, including cholinergic stimuli, inflammatory cytokines, prostaglandins, lipopoly-saccharide, bile salts, nucleotides, nitric oxide, vasoactive intestinal peptide, and neutrophil elastase.93, 95, 103, 113, 124, 125, 126, 127, 128, 129, 130 and 131 Stimulated mucin release can occur immediately and is accompanied by hydration, resulting in approximately a hundredfold expansion in volume of the secretory granule contents.132, 133 Shedding of the large extracellular α-subunits of cell-surface mucins from the cell surface and release of secreted splice variants of cell-surface mucins are less well understood. The protease ADAM17 (also known as TACE) has been shown to trigger shedding of MUC1 in endometrial cells in response to tumor necrosis factor-α,28, 134 and the matrix metalloproteinase-1 also appears to be an effective MUC1 sheddase.29
In addition to regulation of their synthesis and release, mucins are regulated in terms of their glycosylation. Altering mucin carbo-hydrates may block mechanisms that pathogens use to subvert the mucin barrier. Tumor necrosis factor-α alters sialylation of mucins produced by a tracheal cell line135 and expression of both fucosyltransferases and α-2,3-sialyltransferases by normal bronchial mucosal explants.136 In respiratory epithelial cells, the Th2 cytokines IL-4 and IL-13 increase expression of core 2 β-1,6-N-acetylglucosaminyltransferase, which forms β-1,6-branched structures, including core 2, core 4, and blood group I antigen.137 In addition, glycosylation changes occur during infection/inflammation, for example, in individuals with cystic fibrosis or chronic bronchitis,138 as well as H. pylori-infected individuals.69, 139 The inflammation-associated mucin sialylation shown in patients with H. pylori infection returns to the normal pattern following successful bacterial clearance with anti-biotics.140 In rhesus monkeys that share strong similarities in mucin glycosylation and the natural history of H. pylori infection with humans,141H. pylori infection induces time-dependent changes of mucosal glycosylation that alter the H. pylori adhesion targets.69 Such fine-tuned kinetics of host glycosylation dynamically modulate host–bacterial interactions, appearing to balance the impact of infection and thereby may determine the severity of disease.69 Another example of dynamic changes in mucins occurs following infection of rats with the intestinal parasite Nippostrongylus brasiliensis; infection induces increased production and several alterations in the glycosylation of intestinal Muc2 gel-forming mucin, one of which coincides with expulsion of the parasite.142, 143, 144, 145, 146 and 147 These alterations in glycosylation appear to be driven partly by CD4+ T cells, as CD4 but not CD8 depletion blocks the increase in mucin production, change in glycosylation and worm expulsion,148 and also by T-cell-independent mechanisms.149 Such changes in mucin glycosylation need to be considered as a component of innate and adaptive immune responses to mucosal infection.
Microbial Adherence to the Epithelium
To colonize mucosal surfaces and invade the host, microbes typically must first penetrate the secreted mucus barrier and then either attach to the apical surface of epithelial cells decorated with the cell-surface mucins or release toxins that disrupt epithelial integrity. Bacterial adhesion to host cells can be mediated by hydrophobic interactions, cation bridging (i.e., divalent cations counteracting the repulsion of the negatively charged surfaces of bacteria and host) and receptor ligand binding. One of the most extensively studied mechanisms of bacterial adhesion is via lectins and their corresponding glycosylated receptors. Binding is usually of low affinity, but clustering of adhesins and receptors results in multivalent binding. Fimbriae (or pili), outer membrane proteins, and cell wall components (e.g., lipopoly-saccharide) may all function as adhesins. Adhesion can affect the bacteria by stimulation/inhibition of growth as well as induction of other adhesive structures and proteins required for invasion such as secretion systems. On the other hand, effects of adhesion on host cells can include altered morphology, fluid loss, induction of cytokine release, upregulation of adhesion molecules, and apoptosis.150
Many bacterial adhesins bind oligosaccharides present on mucins. Whether bacterial–mucin binding events favor the bacteria or the host is a key question. In reality, for some organisms, this may be a truly commensal relationship with benefits for both the bacteria (by facilitating retention in a favorable niche and even by providing mucin oligosaccharides for meta-bolism) and the host (by retaining bacteria in the outer areas of the mucus barrier where they cannot harm the underlying epithelium and also limiting the niche available for pathogenic bacteria). Numerous interactions between microorganisms and mucins and/or mucin-type carbohydrates have been demonstrated (see Table 3). Bacteria may have multiple adhesins with different carbohydrate specificities, and modulation of surface receptor density, kinetic parameters, or topographical distributions of these receptors on cell membranes regulate adhesion. As an example, H. pylori binds to mucin oligosaccharides via at least four adhesins, which differ substantially with anatomical site along the oro-gastric infection route, mucin type, pH, and gastric disease status.139, 151, 152 and 153 Thus, for H. pylori, binding to mucins can have differing consequences during colonization of the oral-to-gastric niches and during long-term infection.
Mucins as Decoys for Microbial Adhesins
Mucus hypersecretion ensuing from infection is testament to the role of mucus as a component of host defense. Although mucins are the major macromolecular constituent of mucus and are largely responsible for formation of the mucus gel, the precise nature of their role in host defense has not been well demons-trated empirically. Formation of the mucus gel is important in itself, as it provides a biophysical barrier as well as a matrix supporting the retention of a host of antimicrobial molecules. However, the secreted mucins themselves are likely to function as decoys for adhesins that have been evolved by pathogens to engage the cell surface, as the mucins express many of the oligosaccharide structures found on the cell surface and are constitutively produced in large amounts, constantly washing the mucosal surfaces (Figure 1). Some mucins are effective viral agglutinating agents and exogenously applied mucins are effective inhibitors of viral infection in in vitro-cultured cells.154, 155Streptococcus pyogenes, Tritrichomonas foetus, Tritrichomonas mobilensis, influenza viruses, reoviruses, adenoviruses, enteroviruses, and coronaviruses bind to sialic acids, which are present both at the epithelial surface and on mucins.156, 157, 158, 159, 160, 161, 162, 163, 164 and 165
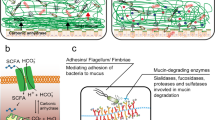
Diagrammatic representation of mucins in the mucosal barrier to infection. (a) The normal mucosa is covered with a continuously replenished thick mucus layer retaining host-defensive molecules. Commensal and environmental microbes may live in the outer mucus layer but the layer ensures that contact of microbes with epithelial cells is rare. (b) Early in infection, many pathogens actively disrupt the mucus layer and thereby gain access to the epithelial cell surface. In addition, this alters the environment for commensal and environmental microbes and opportunistic pathogenesis may occur. (c) Pathogens that break the secreted mucus barrier reach the apical membrane surface, which is decorated with a dense network of large cell-surface mucins. Pathogens bind the cell-surface mucins via lectin interactions and the mucin extracellular domains are shed as releasable decoy molecules. Consequent to contact with microbes and shedding of the extracellular domain, signal transduction by the cytoplasmic domains of the cell-surface mucins modulates cellular response to the presence of microbes. (d) In response to infection, there are alterations in mucins that are driven directly by epithelial cells and in response to signals from underlying innate and adaptive immunity. These alterations include goblet cell hyperplasia and increased mucin secretion and altered mucin glycosylation (depicted by the color change) affecting microbial adhesion and the ability of microbes to degrade mucus. These changes in mucins work in concert with other arms of immunity to clear the infection.
Despite the accepted dogma that secreted mucins limit infection, there are few empirical in vivo data demonstrating their importance. The only secreted mucin for which genetically deficient animals are available is the intestinal mucin, Muc2. Muc2−/− mice develop spontaneous inflammation, presumably due to the absence of the major component of intestinal mucus, leading to increased exposure to the normal intestinal microbial flora.166, 167 As yet, there are no reports of controlled infection experiments in these mice. Further models of secreted mucin deficiency are required to comprehensively determine the importance of secreted mucins in preventing and clearing mucosal infection.
Many pathogens require direct binding to, or penetration of, mucosal epithelial cells to cause pathology. The widest diversity of cell-surface mucin expression is in the mucosal tissues most at risk of infection, such as the gastrointestinal tract, respiratory tract, and eye; notably, nine of the ten cell-surface mucins are expressed in the large intestine, which is the most microbe-rich mucosal environment. Importantly, their ability to be shed from the cell surface has led us to hypothesize that one of the main functions of cell-surface mucins is to act as releasable decoy ligands for microbes attempting to anchor themselves to the glycocalyx. Cell-surface mucins initiate intracellular signaling in response to bacteria, suggesting that they have both a barrier and reporting function on the apical surface of all mucosal epithelial cells.57 However, until recently, much of the evidence had been circumstantial or restricted to in vitro analysis. For example, upregulation of MUC3 expression in colonic cells has been correlated with decreased binding of enteropathogenic E. coli.121, 122In vitro studies have shown that expression of MUC1 by transfection inhibits reovirus and adenovirus infection of MDCK cells by up to tenfold.168, 169 Milk can limit bacterial and viral infections of the gastrointestinal tract and this has been attributed in part to the presence of large amounts of cell-surface mucins, chiefly MUC1 and MUC15, in the milk-fat globule membrane.170, 171 and 172Muc1−/− mice were reported to display chronic infection and inflammation of the reproductive tract, reducing fertility rates. In this latter study, only normal endogenous bacteria were isolated, suggesting that these species become opportunistic pathogens in the absence of Muc1.173 In addition, Muc1−/− mice were reported to have a high frequency of eye inflammation/infection involving Corynebacteria, Staphylococci and Streptococci,174 although this could not be duplicated in a different mouse background held in alternative housing conditions.175
We recently demonstrated that the intestinal pathogen C. jejuni binds to fucosylated mucin oligosaccharides. Controlled infection experiments demonstrated rapid transit of C. jejuni across the gastrointestinal barrier and greater intestinal patho-logy in Muc1−/− mice.55 Bone marrow transplantation studies demonstrated that the increased susceptibility was due to loss of Muc1 on epithelium rather than on leukocytes (which can also express Muc1). Loss of Muc1 had no discernable effects on the abundance or constituency of the intestinal microbial flora. Muc1 appears to prevent C. jejuni infection both by protecting cells from the effects of the cytolethal distending toxin (see above) and by acting as a releasable decoy.55 We have also demonstrated that even though H. pylori can bind Muc1, that primary murine gastric epithelial cells expressing Muc1 bind fewer H. pylori than Muc1−/− cells.176 This paradoxical result is explained by Muc1 acting as a releasable decoy, i.e., the bacteria bind Muc1 expressed on epithelial cells, which is then shed by the host. Due to the absence of this decoy molecule, Muc1−/− mice develop an approximately fivefold greater colonization density of H. pylori from the first days following infection that is maintained for at least 2 months. Consequently, Muc1−/− mice develop severe gastritis not found in wild-type mice.176 Heterozygous mice that have a lower level of gastric Muc1 protein expression show intermediate colonization densities, which suggests that polymorphisms in MUC1 or genes that regulate its expression could underlie susceptibility to H. pylori-induced pathology in human populations. In fact, in humans, individuals with short MUC1 alleles (encoding smaller extra-cellular mucin domains) have a higher propensity to develop gastritis following H. pylori infection.177, 178 and 179 This may be indicative of lower efficacy of smaller MUC1 extracellular mucin domains allowing increased access of bacteria to the epithelial surface, or these alleles may be surrogate markers of polymorphisms influencing the level of gastric MUC1 expression. Recently, a similar protective role has been demonstrated for the extremely large MUC16 cell-surface mucin in human corneal epithelial cells. Greater binding of Staphlylococcus aureus occurs to in vitro-cultured corneal cells when MUC16 is depleted by RNAi.180
Paradoxically, our demonstrations of Muc1-limiting infection in the gastrointestinal tract are opposite to that found in a model of respiratory Pseudomonas aeruginosa infection in the lung in which Muc1−/− mice have an increased clearance of bacteria and a reduced inflammatory response to infection.181 MUC1 binds the P. aeruginosa flagellin,182, 183 but intriguingly appears to inhibit flagellin-stimulated TLR5-mediated activation of NF-κB by an as yet unclear mechanism requiring the MUC1 cytoplasmic domain.181, 184 Whereas an infection-promoting role of a molecule highly expressed on the apical surface of a broad array of mucosal epithelia appears counterintuitive, an anti-inflammatory role for MUC1 in some tissues is consistent with evolutionary adaptations to clear infection by local defense without potentially damaging inflammation, where possible. Further investigations are required with a broad array of pathogens in multiple tissues to clearly delineate the participation of the family of cell-surface mucins in mucosal defense.
Other Protective Roles of Mucins
Mucins have direct and indirect roles in defense from infection distinct from their ability to form a physical barrier and act as adhesion decoys. Not only do mucin oligosaccharides bind microbes, but also, in some cases, they either have direct antimicrobial activity or carry other antimicrobial molecules. A mucin oligosaccharide, α-1-4-linked N-acetyl-glucosamine, which is expressed by some gastric mucins, has been shown to directly interfere with synthesis of H. pylori cell wall components.185 H. pylori must live within gastric mucus to remain protected from luminal gastric pH and prevent expulsion into the intestine. The antimicrobial mucin oligosaccharide probably acts to limit H. pylori expansion within gastric mucus. The non-oligomerizing secreted salivary mucin MUC7 has inherent direct candidacidal activity via a histatin domain at its N-terminus.186, 187 In addition, there is evidence for direct binding of antimicrobial molecules such as histatins and statherin by mucins that would help retain the antimicrobial molecules in the correct mucosal microenvironment where they can best protect the host. For example, MUC7 binds statherin and histatin-1,188 and the other major mucin in saliva, MUC5B, binds histatin-1, -3, and -5 and statherin.189 Secretory IgA (sIgA) is secreted via mucosal epithelial cells and needs to be retained in the immediate mucosal environment to maximize exclusion of pathogens. sIgA is retained at high concentrations in mucus where it can efficiently trap the progress of pathogens, although the mechanism(s) for retaining sIgA in mucus are not well understood. Interestingly, secretory component, which is tightly bound to the Fc region of dimeric-IgA to form sIgA, carries oligosaccharide structures similar to those on mucins.190 In the absence of secretory component carbohydrates, sIgA fails to associate with mucus and fails to prevent infection in a murine respiratory bacterial infection model, substantiating both the physiological importance of sIgA–mucin interactions and the importance of secretory component carbohydrates in maintaining this interaction.191 It is also tempting to speculate that the poly-anionic mucins bind the poly-cationic antimicrobial defensin peptides that are co-secreted into mucus. Interactions of mucins with other secreted antimicrobial molecules has not been fully explored largely due to difficulties in extracting and purifying mucins in the absence of denaturing agents likely to disrupt such interactions. The cell-surface mucins are an integral component of the glycocalyx where they are likely to interact with proteoglycans and other molecules that could retain host defense molecules in a molecular complex covering the apical cell surface.192, 193 Therefore, other mucins and mucin oligosaccharides may yet prove to have direct and indirect antimicrobial activity. Regardless of whether antimicrobial molecules are retained in mucus by direct binding with mucins or by the biophysical properties of mucus, if mucin synthesis is aberrant or secreted mucins are degraded, the antimicrobial molecules will have impaired efficacy.
Subversion of the Mucin Barrier by Mucosal Pathogens
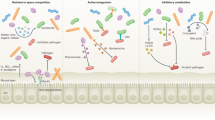
Perhaps the best evidence for the importance of the mucin barrier to infection is the wide variety of strategies used by microbes to subvert or avoid this barrier. Mucin barrier subversion strategies used by microbes include the production of enzymes capable of degrading mucin core proteins and mucin carbohydrates, and effective motility through mucus gels. Motility is important for bacterial mucosal pathogens to facilitate breaking through the physical mucus barrier. In fact, a vast proportion of mucosal bacterial pathogens are flagellated.194, 195H. pylori that have dysfunctional flagella have a greatly reduced ability to infect.196H. pylori uses motility for initial colonization and to attain robust infection. In conjunction with motility, degradative enzymes such as glycosulfatases, sialidases, sialate O-acetylesterases, N-acetyl neuraminate lyases and mucinases are produced by a broad range of bacterial pathogens to destabilize the mucus gel and remove mucin decoy carbohydrates for adhesins.197, 198, 199, 200 and 201 The protozoan parasite Entamoeba histolytica cleaves the MUC2 mucin, which is the major structural component of the intestinal mucus, and this cleavage is predicted to depolymerize the MUC2 polymers.202 The size of the polymer is important for the formation of entangled gels and the viscous properties of mucus; consequently, cleavage of the mucin polymer will effectively result in a local disintegration of mucus.203 There is evidence that these degradative enzymes are critical for microbial pathogenesis. For example, the Vibrio cholerae Hap A, which has both mucinolytic and cytotoxic activity, is induced by mucin and required for translocation through mucin-containing gels.204 The widespread and critically required expression of neuraminidases by a wide variety of sialic acid-binding mucosal viruses underlines the importance of elimination of mucin carbohydrates for their pathogenicity.160 Lipopolysaccharide from H. pylori decreases mucin synthesis,123 and the mucin carbohydrate-binding adhesins BabA and SabA undergo phase variation and change expression during infection,153, 205 which may allow them to evade this host defense mechanism.
Avoidance of the Mucin Barrier by Mucosal Pathogens
Another strategy commonly used by mucosal pathogens is to avoid the mucin barrier. Intestinal M cells, specifically designed to capture and present microbes to the underlying lymphoid tissue, can be regarded as a hole in the mucin barrier. The dome epithelium in which they lie lacks goblet cells, and therefore does not produce gel-forming mucins, and their apical cell surface has only sparse microvilli and an apparently thin glycocalyx.206, 207 Although no studies have measured the expression of individual cell-surface mucins in M cells, there appear to be differences in the glycocalyx mucins between M cells and adjacent intestinal mucosal epithelial cells. In some species, M cells can be identified by their pattern of lectin binding to specific cell-surface carbohydrates that differ with other mucosal epithelial cells.208, 209 Consequently, even though M cells constitute only a very small percentage of mucosal epithelial cells, they are the major point of attachment and/or entry used by a large number of mucosal pathogens including bacteria (e.g., S. typhimurium, Shigella flexneri, Yersinia enterocolitica, and V. cholerae), viruses (e.g., reovirus, HIV-1, and polio virus) and parasites (e.g., Cryptosporidia).206, 210 and 211 Another strategy used by pathogens to avoid the cell-surface mucin barrier, once mucus is penetrated or M cells are invaded, is to disrupt the tight junctions between adjacent mucosal epithelial cells thereby exposing the vulnerable lateral membranes not protected by the glycocalyx. Such examples include S. flexneri,212 enteropathogenic E. coli,213Porphyromonas gingivalis214 and H. pylori.215
Models to Investigate Interactions between Microbes and Mucins
Numerous models, including cancer cell-lines, organ cultures of gastric biopsies and whole animals have been used to investigate mucin–microbe interactions. Although they express orthologs to most human mucins, the most commonly used laboratory animals such as rats and mice have differing glycosylation of some of their mucins. In fact, it is tempting to speculate that differences in mucin glycosylation between mammalian species may underlie some of the differences in infectivity/pathogenicity for individual microbial pathogens. Murine knockout models are only available for Muc1,216Muc2,166 and Muc13 (M.A. McGuckin, unpublished data), and there are also mutants with aberrant Muc2 assembly.217 Thus, there is a need for more models, as mouse knockouts, although limited by the slightly different glycosylation, still represent an important way to collect information of the in vivo function of mucins in infection. Because human pathogens commonly have adhesins for human carbohydrate structures, it is important to select appropriate models for individual pathogens. For example, the effects of H. pylori infection on the mouse are mild, and gastric cancer is not induced even after long-term exposure without other stimuli or genetic defects, although the mouse may develop chronic atrophic gastritis.218, 219H. pylori can colonize the guinea pig and the Mongolian gerbil and cause a severe inflammatory response but does not induce cancer in the absence of exogenous chemical carcinogens.220 These small animal models are useful to study some aspects of H. pylori infection and have the advantage of being relatively cheap. In contrast, rhesus monkeys naturally have persistent H. pylori infection leading to loss of mucus, gastritis, gastric ulcers and even cancer.221, 222, 223 and 224 In addition, the anatomy and physiology of the GI tract of the rhesus monkey, as well as the expression of mucins and mucin glycosylation are very similar to that in human.141 However, this model is expensive, the monkeys can have preexisting natural infection, and primate research has a higher level of ethical considerations.
In vitro microbial–mammalian cocultures are used extensively to elucidate the mechanisms by which microbes adhere, invade, and signal to the host, and to examine ensuing mammalian cell responses. These complex interactions are reliant on appropriate gene expression and cellular functioning of both the microbial and mammalian cells. It is therefore critical that appropriate microbial and mammalian cells are used and that the environment created experimentally is as similar to the human mucosal environment as possible. Human cell lines commonly used for in vitro infection studies have a highly variable expression of mucins and mucin glycosylation, and generally have very low production and secretion of gel-forming mucins.225 Investigators using these models need to be aware of these limitations and consider them in interpreting their data. Additional important issues to consider are choice of cell line and, depending on the type of bacteria, oxygen tension.225, 226 With respect to appropriate mucin production, primary human tracheobronchial epithelial cells cultured in an air–liquid interface represent the most physiological cell cultures in which infection studies can currently be undertaken.227Ex vivo-cultured tissue explants provide another potential avenue for exploring microbial mucin interactions in vitro.228, 229 and 230
Conclusions
The personal repertoire of expression of mucin core proteins and their glycans, mucin allele length, and transient changes in mucin expression and glycosylation in response to infection or stress, as well as variations in environmental conditions may all affect microbial interaction with host mucins and the pathogenic consequences of microbial colonization. Rather than a static barrier, mucins should be considered as a dynamic responsive component of the mucosal barrier that interacts with and responds to other elements of innate and adaptive immunity. Difficulties in working with these complex glycoproteins and the paucity of physiological experimental systems need to be overcome if we are to fully understand the roles of mucins in host defense from infection.
Disclosure
The authors declare no conflict of interest.
References
Kagnoff, M.F. & Eckmann, L. Epithelial cells as sensors for microbial infection. J. Clin. Invest. 100, 6–10 (1997).
Lu, J., Teh, C., Kishore, U. & Reid, K.B. Collectins and ficolins: sugar pattern recognition molecules of the mammalian innate immune system. Biochim. Biophys. Acta 1572, 387–400 (2002).
Raj, P.A. & Dentino, A.R. Current status of defensins and their role in innate and adaptive immunity. FEMS Microbiol. Lett. 206, 9–18 (2002).
Prydal, J.I., Muir, M.G. & Dilly, P.N. Comparison of tear film thickness in three species determined by the glass fibre method and confocal microscopy. Eye 7, 472–475 (1993).
Mercer RR ML, R. & Crapo, J.D. Mucous lining layers in human and rat airways. Ann. Rev. Resp. Dis. 145, 355 (1992).
Strugala, V., Allen, A., Dettmar, P.W. & Pearson, J.P. Colonic mucin: methods of measuring mucus thickness. Proc. Nutr. Soc. 62, 237–243 (2003).
Jordan, N., Newton, J., Pearson, J. & Allen, A. A novel method for the visualization of the in situ mucus layer in rat and man. Clin. Sci. (London) 95, 97–106 (1998).
Atuma, C., Strugala, V., Allen, A. & Holm, L. The adherent gastrointestinal mucus gel layer: thickness and physical state in vivo. Am. J. Physiol. Gastrointest. Liver Physiol. 280, G922–G929 (2001).
Soler, M. et al. Adhesion-related glycocalyx study: quantitative approach with imaging-spectrum in the energy filtering transmission electron microscope (EFTEM). FEBS Lett. 429, 89–94 (1998).
Ito, S. Structure and function of the glycocalyx. Fed. Proc. 28, 12–25 (1969).
Madara, J. & Trier, J. Functional morphology of the mucosa of the small intestine. In Physiology of the Gastrointestinal Tract (Johnson L.R., ed) 1269–1299 (Raven press, 1987 ).
Klein, A. et al. Isolation and structural characterization of novel sialylated oligosaccharide-alditols from respiratory-mucus glycoproteins of a patient suffering from bronchiectasis. Eur. J. Biochem. 211, 491–500 (1993).
Pigny, P. et al. Human mucin genes assigned to 11p15.5: identification and organization of a cluster of genes. Genomics 38, 340–352 (1996).
Desseyn, J.L., Aubert, J.P., Porchet, N. & Laine, A. Evolution of the large secreted gel-forming mucins. Mol. Biol. Evol. 17, 1175–1184 (2000).
Desseyn, J.L. et al. Evolutionary history of the 11p15 human mucin gene family. J. Mol. Evol. 46, 102–106 (1998).
Chen, Y. et al. Genome-wide search and identification of a novel gel-forming Mucin MUC19/Muc19 in glandular tissues. Am. J. Respir. Cell Mol. Biol. 30, 155–165 (2003).
Gum, J.R., Hicks, J.W., Toribara, N.W., Siddiki, B. & Kim, Y.S. Molecular cloning of human intestinal mucin (MUC2) cDNA. Identification of the amino terminus and overall sequence similarity to prepro-von Willebrand factor. J. Biol. Chem. 269, 2440–2446 (1994).
Perez-Vilar, J., Eckhardt, A.E., DeLuca, A. & Hill, R.L. Porcine submaxillary mucin forms disulfide-linked multimers through its amino-terminal d-domains. J. Biol. Chem. 273, 14442–14449 (1998).
Asker, N., Baeckstrom, D., Axelsson, M.A.B., Carlstedt, I. & Hansson, G.C. The human MUC2 mucin apoprotein appears to dimerize before O-glycosylation and shares epitopes with the insoluble mucin of rat small intestine. Biochem. J. 308, 873–880 (1995).
Perez-Vilar, J. & Hill, R.L. The structure and assembly of secreted mucins. J. Biol. Chem. 274, 31751–31754 (1999).
Lidell, M.E. et al. The recombinant C-terminus of the human MUC2 mucin forms dimers in Chinese-hamster ovary cells and heterodimers with full-length MUC2 in LS 174 T cells. Biochem. J. 372, 335–345 (2003).
Godl, K. et al. The N terminus of the MUC2 mucin forms trimers that are held together within a trypsin-resistant core fragment. J. Biol. Chem. 277, 47248–47256 (2002).
Sheehan, J.K. et al. Physical characterization of the MUC5AC mucin: a highly oligomeric glycoprotein whether isolated from cell culture or in vivo from respiratory mucous secretions. Biochem. J. 347 (Part 1), 37–44 (2000).
Sheehan, J.K., Howard, M., Richardson, P.S., Longwill, T. & Thornton, D.J. Physical characterization of a low-charge glycoform of the MUC5B mucin comprising the gel-phase of an asthmatic respiratory mucous plug. Biochem. J. 338, 507–513 (1999).
Jentoft, N. Why are proteins O-glycosylated? Trends Biochem. Sci. 15, 291–294 (1990).
Wreschner, D.H. et al. Generation of ligand-receptor alliances by “SEA” module-mediated cleavage of membrane-associated mucin proteins. Protein Sci. 11, 698–706 (2002).
Macao, B., Johansson, D.G., Hansson, G.C. & Hard, T. Autoproteolysis coupled to protein folding in the SEA domain of the membrane-bound MUC1 mucin. Nat. Struct. Mol. Biol. 13, 71–76 (2006).
Thathiah, A., Blobel, C.P. & Carson, D.D. Tumor necrosis factor-alpha converting enzyme/ADAM 17 mediates MUC1 shedding. J. Biol. Chem. 278, 3386–3394 (2003).
Thathiah, A. & Carson, D.D. MT1-MMP mediates MUC1 shedding independently of TACE/ADAM17. Biochem. J. 382 (Part 1), 363–373 (2004).
Lillehoj, E.P., Han, F. & Kim, K.C. Mutagenesis of a Gly-Ser cleavage site in MUC1 inhibits ectodomain shedding. Biochem. Biophys. Res. Commun. 307, 743–749 (2003).
Zrihan-Licht, S. et al. Characterization and molecular cloning of a novel MUC1 protein devoid of tandem repeats expressed in human breast cancer tissue. Eur. J. Biochem. 224, 787–795 (1994).
Williams, S.J. et al. Two novel mucin genes downregulated in colorectal cancer identified by differential display. Cancer Res. 59, 4083–4089 (1999).
Williams, S.J., Munster, D.J., Quin, R.J., Gotley, D.C. & McGuckin, M.A. The MUC3 gene encodes a transmembrane mucin and is alternatively spliced. Biochem. Biophys. Res. Commun. 261, 83–89 (1999).
Choudhury, A. et al. Alternate splicing at the 3′-end of the human pancreatic tumor-associated mucin MUC4 cDNA. Teratog. Carcinog. Mutagen 21, 83–96 (2001).
Williams, S.J. et al. Muc13, a novel human cell surface mucin expressed by epithelial and hemopoietic cells. J. Biol. Chem. 276, 18327–18336 (2001).
Hilkens, J., Ligtenberg, M.J., Vos, H.L. & Litvinov, S.V. Cell membrane-associated mucins and their adhesion-modulating property. Trends Biochem. Sci. 17, 359–363 (1992).
Litvinov, S.V. & Hilkens, J. The epithelial sialomucin, episialin, is sialylated during recycling. J. Biol. Chem. 268, 21364–21371 (1993).
Altschuler, Y. et al. Clathrin-mediated endocytosis of MUC1 is modulated by its glycosylation state. Mol. Biol. Cell 11, 819–831 (2000).
Thingstad, T., Vos, H.L. & Hilkens, J. Biosynthesis and shedding of epiglycanin: a mucin-type glycoprotein of the mouse TA3Ha mammary carcinoma cell. Biochem. J. 353, 33–40 (2001).
Pandey, P., Kharbanda, S. & Kufe, D. Association of the DF3/MUC1 breast cancer antigen with Grb2 and the Sos/Ras exchange protein. Cancer Res. 55, 4000–4003 (1995).
Li, Y., Kuwahara, H., Ren, J., Wen, G. & Kufe, D. The c-Src tyrosine kinase regulates signaling of the human DF3/MUC1 carcinoma-associated antigen with GSK3 beta and beta-catenin. J. Biol. Chem. 276, 6061–6064 (2001).
Li, Q., Ren, J. & Kufe, D. Interaction of human MUC1 and beta-catenin is regulated by Lck and ZAP-70 in activated Jurkat T cells. Biochem. Biophys. Res. Commun. 315, 471–476 (2004).
Regimbald, L.H. et al. The breast mucin MUC1 as a novel adhesion ligand for endothelial intercellular adhesion molecule 1 in breast cancer. Cancer Res. 56, 4244–4249 (1996).
Nath, D. et al. Macrophage-tumour cell interactions: identification of muc1 on breast cancer cells as a potential counter-receptor for the macrophage-restricted receptor, sialoadhesin. Immunology 98, 213–219 (1999).
Gubbels, J.A. et al. Mesothelin-MUC16 binding is a high affinity, N-glycan dependent interaction that facilitates peritoneal metastasis of ovarian tumors. Mol. Cancer 5, 50 (2006).
Ciborowski, P. & Finn, O.J. Non-glycosylated tandem repeats of MUC1 facilitate attachment of breast tumor cells to normal human lung tissue and immobilized extracellular matrix proteins (ECM) in vitro: potential role in metastasis. Clin. Exp. Metastasis 19, 339–345 (2002).
Komatsu, M., Carraway, C.A.C., Fregien, N.L. & Carraway, K.L. Reversible disruption of cell-matrix and cell–cell interactions by overexpression of sialomucin complex. J. Biol. Chem. 272, 33245–33254 (1997).
Li, Y.Q., Bharti, A., Chen, D.S., Gong, J.L. & Kufe, D. Interaction of glycogen synthase kinase 3β with the DF3/MUC1 carcinoma-associated antigen and β-catenin. Mol. Cell Biol. 18, 7216–7224 (1998).
Quin, R.J. & McGuckin, M.A. Phosphorylation of MUC1 correlates with changes in cell–cell adhesion. Int. J. Cancer 87, 499–506 (2000).
Li, Y. et al. The epidermal growth factor receptor regulates interaction of the human DF3/MUC1 carcinoma antigen with c-Src and beta-catenin. J. Biol. Chem. 276, 35239–35242 (2001).
Ren, J., Li, Y. & Kufe, D. Protein kinase C delta regulates function of the DF3/MUC1 carcinoma antigen in beta-catenin signaling. J. Biol. Chem. 277, 17616–17622 (2002).
Li, Y., Liu, D., Chen, D., Kharbanda, S. & Kufe, D. Human DF3/MUC1 carcinoma-associated protein functions as an oncogene. Oncogene 22, 6107–6110 (2003).
Ren, J. et al. Human MUC1 carcinoma-associated protein confers resistance to genotoxic anticancer agents. Cancer Cell 5, 163–175 (2004).
Wei, X., Xu, H. & Kufe, D. Human MUC1 oncoprotein regulates p53-responsive gene transcription in the genotoxic stress response. Cancer Cell 7, 167–178 (2005).
McAuley, J.L. et al. MUC1 cell surface mucin is a critical element of the mucosal barrier to infection. J. Clin. Invest. 117, 2313–2324 (2007).
Lara-Tejero, M. & Galan, J.E. A bacterial toxin that controls cell cycle progression as a deoxyribonuclease I-like protein. Science 290, 354–357 (2000).
Lillehoj, E.P., Kim, H., Chun, E.Y. & Kim, K.C. Pseudomonas aeruginosa stimulates phosphorylation of the airway epithelial membrane glycoprotein Muc1 and activates MAP kinase. Am. J. Physiol. Lung Cell Mol. Physiol. 287, L809–L815 (2004).
McCool, D.J., Okada, Y., Forstner, J.F. & Forstner, G.G. Roles of calreticulin and calnexin during mucin synthesis in LS180 and HT29/A1 human colonic adenocarcinoma cells. Biochem. J. 341 (Part 3), 593–600 (1999).
Ho, J.J. et al. N-glycosylation is required for the surface localization of MUC17 mucin. Int. J. Oncol. 23, 585–592 (2003).
Kui Wong, N. et al. Characterization of the oligosaccharides associated with the human ovarian tumor marker CA125. J. Biol. Chem. 278, 28619–28634 (2003).
Oriol, R. ABO, Hh, Lewis, and secretion serology, genetics, and tissue distribution. In Blood Cell Biochemistry, Vol 6 (Cartron, J., Rouger, P., eds) 37–73 (Plenum press, New York, 1995 ).
Oriol, R., Le Pendu, J. & Mollicone, R. Genetics of ABO, H, Lewis, X and related antigens. Vox Sang 51, 161–171 (1986).
Henry, S., Oriol, R. & Samuelsson, B. Lewis histo-blood group system and associated secretory phenotypes. Vox Sang 69, 166–182 (1995).
Yu, L.C. et al. Polymorphism and distribution of the secretor alpha(1,2)-fucosyltransferase gene in various Taiwanese populations. Transfusion 41, 1279–1284 (2001).
Gerken, T.A. Kinetic modeling confirms the biosynthesis of mucin core 1 (beta-Gal(1-3) alpha-GalNAc-O-Ser/Thr) O-glycan structures are modulated by neighboring glycosylation effects. Biochemistry 43, 4137–4142 (2004).
Marionneau, S. et al. ABH and Lewis histo-blood group antigens, a model for the meaning of oligosaccharide diversity in the face of a changing world. Biochimie 83, 565–573 (2001).
Marionneau, S., Airaud, F., Bovin, N.V., Le Pendu, J. & Ruvoen-Clouet, N. Influence of the combined ABO FUT2, and FUT3 polymorphism on susceptibility to Norwalk virus attachment. J. Infect. Dis. 192, 1071–1077 (2005).
Lomberg, H., Jodal, U., Leffler, H., De Man, P. & Svanborg, C. Blood group non-secretors have an increased inflammatory response to urinary tract infection. Scand. J. Infect. Dis. 24, 77–83 (1992).
Linden, S.K. et al. Glycan secretor phenotypes modulate mucosal innate immunity and affect H. pylori infection. PLoS Pathog. 4, e2 (2008).
Aspholm-Hurtig, M. et al. Functional adaptation of BabA, the H. pylori ABO blood group antigen binding adhesin. Science 305, 519–522 (2004).
Rogers, D.F. Airway goblet cells: responsive and adaptable front-line defenders. Eur. Respir. J. 7, 1690–1706 (1994).
Kovarik, A., Peat, N., Wilson, D., Gendler, S.J. & Taylor-Papadimitriou, J. Analysis of the tissue-specific promoter of the MUC1 gene. J. Biol. Chem. 268, 9917–9926 (1993).
Abe, M. & Kufe, D. Transcriptional regulation of DF3 gene expression in human MCF-7 breast carcinoma cells. J. Cell Physiol. 143, 226–231 (1990).
Zaretsky, J.Z. et al. Analysis of the promoter of the MUC1 gene overexpressed in breast cancer. FEBS Lett. 461, 189–195 (1999).
Velcich, A., Palumbo, L., Selleri, L., Evans, G. & Augenlicht, L. Organization and regulatory aspects of the human intestinal mucin gene (MUC2) locus. J. Biol. Chem. 272, 7968–7976 (1997).
Gum, J.R., Hicks, J.W. & Kim, Y.S. Identification and characterization of the MUC2 (human intestinal mucin) gene 5′-flanking region: promoter activity in cultured cells. Biochem. J. 325, 259–267 (1997).
Nogami, H. et al. Sp1 protein contributes to airway specific rat Muc2 mucin gene transcription. Gene 198, 191–201 (1997).
Gum, J.R. Jr et al. Goblet cell-specific expression mediated by the MUC2 mucin gene promoter in the intestine of transgenic mice. Am. J. Physiol. 276, G666–G676 (1999).
Perrais, M., Pigny, P., Copin, M.C., Aubert, J.P. & Van Seuningen, I. Induction of MUC2 and MUC5AC mucin by factors of the epidermal growth factor family is mediated by EGF-R/Ras/Raf/MAPK signaling cascade and Sp1. J. Biol. Chem. 19, 19 (2002).
Yamamoto, H., Bai, Y.Q. & Yuasa, Y. Homeodomain protein CDX2 regulates goblet-specific MUC2 gene expression. Biochem. Biophys. Res. Commun. 300, 813–818 (2003).
van der Sluis, M. et al. The murine Muc2 mucin gene is transcriptionally regulated by the zinc-finger GATA-4 transcription factor in intestinal cells. Biochem. Biophys. Res. Commun. 325, 952–960 (2004).
Shekels, L.L. & Ho, S.B. Characterization of the mouse Muc3 membrane bound intestinal mucin 5′ coding and promoter regions: regulation by inflammatory cytokines. Biochim. Biophys. Acta 1627, 90–100 (2003).
Perez, A., Barco, R., Fernandez, I., Price-Schiavi, S.A. & Carraway, K.L. PEA3 transactivates the Muc4/Sialomucin complex promoter in mammary epithelial and tumor cells. J. Biol. Chem. 278, 36942–36952 (2003).
Fauquette, V. et al. Transcription factor AP-2alpha represses both the Mucin MUC4 expression and pancreatic cancer cell proliferation. Carcinogenesis 28, 2305–2312 (2007).
Perrais, M. et al. Characterization of human mucin gene MUC4 promoter: importance of growth factors and proinflammatory cytokines for its regulation in pancreatic cancer cells. J. Biol. Chem. 276, 30923–30933 (2001).
Kim, S.W. et al. Regulation of mucin gene expression by CREB via a nonclassical RA signaling pathway. Mol. Cell Biol. 27, 6933–6947 (2007).
Van Seuningen, I., Perrais, M., Pigny, P., Porchet, N. & Aubert, J.P. Sequence of the 5′-flanking region and promoter activity of the human mucin gene MUC5B in different phenotypes of colon cancer cells. Biochem. J. 348, 675–686 (2000).
Shirotani, K., Taylor-Papadimitriou, J., Gendler, S.J. & Irimura, T. Transcriptional regulation of the MUC1 mucin gene in colon carcinoma cells by a soluble factor—identification of a regulatory element. J. Biol. Chem. 269, 15030–15035 (1994).
Vos, H.L., van der Valk, S.W., Prinsenberg, T., Mooi, W.J. & Hilkens, J. IL6-induction of the MUC1/episialin promoter in T47D cells. In Proceedings of 4th International Workshop on Carcinoma-Associated Mucins 30 (1996).
Temann, U.A. et al. A novel role for murine IL-4 in vivo: induction of MUC5ac gene expression and mucin hypersecretion. Am. J. Respir. Cell Mol. Biol. 16, 471–478 (1997).
Dabbagh, K. et al. IL-4 induces mucin gene expression and goblet cell metaplasia in vitro and in vivo. J. Immunol. 162, 6233–6237 (1999).
Longphre, M. et al. Allergen-induced IL-9 directly stimulates mucin transcription in respiratory epithelial cells. J. Clin. Invest. 104, 1375–1382 (1999).
Smirnova, M.G., Birchall, J.P. & Pearson, J.P. TNF-alpha in the regulation of MUC5AC secretion: some aspects of cytokine-induced mucin hypersecretion on the in vitro model. Cytokine 12, 1732–1736 (2000).
Kim, Y.D. et al. Regulation of IL-1beta-mediated MUC2 gene in NCI-H292 human airway epithelial cells. Biochem. Biophys. Res. Commun. 274, 112–116 (2000).
Enss, M.L. et al. Proinflammatory cytokines trigger MUC gene expression and mucin release in the intestinal cancer cell line LS180. Inflamm. Res. 49, 162–169 (2000).
Louahed, J. et al. Interleukin-9 upregulates mucus expression in the airways. Am. J. Respir. Cell Mol. Biol. 22, 649–656 (2000).
Shim, J.J. et al. IL-13 induces mucin production by stimulating epidermal growth factor receptors and by activating neutrophils. Am. J. Physiol. Lung Cell Mol. Physiol. 280, L134–L140 (2001).
Gaemers, I.C., Vos, H.L., Volders, H.H., van der Valk, S.W. & Hilkens, J. A stat-responsive element in the promoter of the episialin/MUC1 gene is involved in its overexpression in carcinoma cells. J. Biol. Chem. 276, 6191–6199 (2001).
Smirnova, M.G., Kiselev, S.L., Birchall, J.P. & Pearson, J.P. Up-regulation of mucin secretion in HT29-MTX cells by the pro-inflammatory cytokines tumor necrosis factor-alpha and interleukin-6. Eur. Cytokine Netw. 12, 119–125 (2001).
Whittaker, L. et al. Interleukin-13 mediates a fundamental pathway for airway epithelial mucus induced by CD4 T cells and interleukin-9. Am. J. Respir. Cell Mol. Biol. 27, 593–602 (2002).
Seong, J.K. et al. Upregulation of MUC8 and downregulation of MUC5AC by inflammatory mediators in human nasal polyps and cultured nasal epithelium. Acta Otolaryngol. 122, 401–407 (2002).
Kim, Y.D. et al. Interleukin-1beta induces MUC2 gene expression and mucin secretion via activation of PKC-MEK/ERK, and PI3K in human airway epithelial cells. J. Korean Med. Sci. 17, 765–771 (2002).
Smirnova, M.G., Guo, L., Birchall, J.P. & Pearson, J.P. LPS up-regulates mucin and cytokine mRNA expression and stimulates mucin and cytokine secretion in goblet cells. Cell Immunol. 221, 42–49 (2003).
Iwashita, J. et al. mRNA of MUC2 is stimulated by IL-4, IL-13 or TNF-alpha through a mitogen-activated protein kinase pathway in human colon cancer cells. Immunol. Cell Biol. 81, 275–282 (2003).
Song, K.S. et al. Interleukin-1 beta and tumor necrosis factor-alpha induce MUC5AC overexpression through a mechanism involving ERK/p38 mitogen-activated protein kinases-MSK1-CREB activation in human airway epithelial cells. J. Biol. Chem. 278, 23243–23250 (2003).
Li, S., Intini, G. & Bobek, L.A. Modulation of MUC7 mucin expression by exogenous factors in airway cells in vitro and in vivo. Am. J. Respir. Cell Mol. Biol. 35, 95–102 (2006).
Damera, G., Xia, B. & Sachdev, G.P. IL-4 induced MUC4 enhancement in respiratory epithelial cells in vitro is mediated through JAK-3 selective signaling. Respir. Res. 7, 39 (2006).
Koga, T. et al. TNF-alpha induces MUC1 gene transcription in lung epithelial cells: Its signaling pathway and biological implication. Am. J. Physiol. Lung Cell Mol. Physiol. 293, L693–L701 (2007).
Andrianifahanana, M. et al. IFN-gamma-induced expression of MUC4 in pancreatic cancer cells is mediated by STAT-1 upregulation: a novel mechanism for IFN-gamma response. Oncogene 26, 7251–7261 (2007).
Song, J.S. et al. Nitric oxide induces MUC5AC mucin in respiratory epithelial cells through PKC and ERK dependent pathways. Respir. Res. 8, 28 (2007).
Fischer, B.M. & Voynow, J.A. Neutrophil elastase induces MUC5AC gene expression in airway epithelium via a pathway involving reactive oxygen species. Am. J. Respir. Cell Mol. Biol. 26, 447–452 (2002).
Fischer, B.M. et al. Neutrophil elastase increases MUC4 expression in normal human bronchial epithelial cells. Am. J. Physiol. Lung Cell Mol. Physiol. 284, L671–L679 (2003).
Voynow, J.A. et al. Neutrophil elastase increases MUC5AC mRNA and protein expression in respiratory epithelial cells. Am. J. Physiol. Lung Cell Mol. Physiol. 20, 835–843 (1999).
Lundgren, J.D., Rieves, R.D., Mullol, J., Logun, C. & Shelhamer, J.H. The effect of neutrophil protenase enzymes on the release of mucus from feline and human airway cultures. Resp. Med. 88, 511–518 (1994).
Dwyer, T.M. & Farley, J.M. Human neutrophil elastase releases two pools of mucinlike glycoconjugate from tracheal submucosal gland cells. Am. J. Physiol. Lung Cell Mol. Physiol. 278, L675–L682 (2000).
Kuwahara, I. et al. Neutrophil elastase stimulates MUC1 gene expression through increased Sp1 binding to the MUC1 promoter. Am. J. Physiol. Lung Cell Mol. Physiol. 289, L355–L362 (2005).
Yanagihara, K., Seki, M. & Cheng, P.W. Lipopolysaccharide induces mucus cell metaplasia in mouse lung. Am. J. Respir. Cell Mol. Biol. 24, 66–73 (2001).
Lemjabbar, H. & Basbaum, C. Platelet-activating factor receptor and ADAM10 mediate responses to Staphylococcus aureus in epithelial cells. Nat. Med. 8, 41–46 (2002).
Dohrman, A. et al. Mucin gene (MUC2 and MUC5AC) upregulation by Gram-positive and Gram-negative bacteria. Biochim. Biophys. Acta 1406, 251–259 (1998).
Caballero-Franco, C., Keller, K., De Simone, C. & Chadee, K. The VSL#3 probiotic formula induces mucin gene expression and secretion in colonic epithelial cells. Am. J. Physiol. Gastrointest. Liver Physiol. 292, G315–G322 (2007).
Mack, D.R., Michail, S., Wei, S., McDougall, L. & Hollingsworth, M.A. Probiotics inhibit enteropathogenic E. coli adherence in vitro by inducing intestinal mucin gene expression. Am. J. Physiol. 276, G941–G950 (1999).
Mack, D.R., Ahrne, S., Hyde, L., Wei, S. & Hollingsworth, M.A. Extracellular MUC3 mucin secretion follows adherence of Lactobacillus strains to intestinal epithelial cells in vitro. Gut 52, 827–833 (2003).
Slomiany, B.L. & Slomiany, A. Cytosolic phospholipase A2 activation in Helicobacter pylori lipopolysaccharide-induced interference with gastric mucin synthesis. IUBMB Life 58, 217–223 (2006).
Kim, K.C., Lee, B.C., Pou, S. & Ciccolella, D. Effects of activation of polymorphonuclear leukocytes on airway goblet cell mucin release in a co-culture system. Inflamm. Res. 52, 258–262 (2003).
Fischer, B.M., Krunkosky, T.M., Wright, D.T., Dolanokeefe, M. & Adler, K.B. Tumor necrosis factor-alpha (TNF-alpha) stimulates mucin secretion and gene expression in airway epithelium in vitro. Chest 107 (3 Suppl), 133S–135S (1995).
Hollande, E., Fanjul, M., Claret, S., Forguelafitte, M.E. & Bara, J. Effects of VIP on the regulation of mucin secretion in cultured human pancreatic cancer cells (CAPAN-1). In vitro Cell Dev. Biol. 31, 227–233 (1995).
Kishioka, C., Okamoto, K., Kim, J. & Rubin, B.K. Regulation of secretion from mucous and serous cells in the excised ferret trachea. Respir. Physiol. 126, 163–171 (2001).
Klinkspoor, J.H. et al. Mucin secretion by the human colon cell line LS174T is regulated by bile salts. Glycobiology 9, 13–19 (1999).
Jarry, A., Vallette, G., Branka, J.E. & Laboisse, C. Direct secretory effect of interleukin-1 via type I receptors in human colonic mucous epithelial cells (HT29-C1.16E). Gut 38, 240–242 (1996).
Kim, K.C., Park, H.R., Shin, C.Y., Akiyama, T. & Ko, K.H. Nucleotide-induced mucin release from primary hamster tracheal surface epithelial cells involves the p-2u purinoceptor. Eur. Respir. J. 9, 542–548 (1996).
Gottke, M. & Chadee, K. Exogenous nitric oxide stimulates mucin secretion from LS174T colonic adenocarcinoma cells. Inflamm. Res. 45, 209–212 (1996).
Tam, P.Y. & Verdugo, P. Control of mucus hydration as a Donnan equilibrium process. Nature 292, 340–342 (1981).
Verdugo, P. Mucin exocytosis. Am. Rev. Respir. Dis. 144, S33–S37 (1991).
Thathiah, A. et al. TNF-alpha stimulates MUC1 synthesis and Ectodomain release in a human uterine epithelial cell line. Endocrinology 145, 4192–4203 (2004).
Delmotte, P. et al. Tumor necrosis factor alpha increases the expression of glycosyltransferases and sulfotransferases responsible for the biosynthesis of sialylated and/or sulfated Lewis x epitopes in the human bronchial mucosa. J. Biol. Chem. 277, 424–431 (2002).
Delmotte, P. et al. Influence of TNFalpha on the sialylation of mucins produced by a transformed cell line MM-39 derived from human tracheal gland cells. Glycoconj. J. 18, 487–497 (2001).
Beum, P.V., Basma, H., Bastola, D.R. & Cheng, P.W. Mucin biosynthesis: upregulation of core 2 beta 1,6 N-acetylglucosaminyltransferase by retinoic acid and Th2 cytokines in a human airway epithelial cell line. Am. J. Physiol. Lung Cell Mol. Physiol. 288, L116–L124 (2005).
Schulz, B.L. et al. Glycosylation of sputum mucins is altered in cystic fibrosis patients. Glycobiology 17, 698–712 (2007).
Linden, S.K., Wickstrom, C., Lindell, G. & Carlstedt, I. Four modes of adhesion are used during Helicobacter pylori binding to human mucins in the oral and gastric niches. Helicobacter (in press) (2008).
Ota, H. et al. Helicobacter pylori infection produces reversible glycosylation changes to gastric mucins. Virchows Arch. B Cell Pathol. Incl. Mol. Pathol. 433, 419–426 (1998).
Lindén, S., Borén, T., Dubois, A. & Carlstedt, I. Rhesus monkey gastric mucins and their Helicobacter pylori binding properties. Biochem. J. 379, 1–11 (2004).
Koninkx, J.F., Mirck, M.H., Hendriks, H.G., Mouwen, J.M. & van Dijk, J.E. Nippostrongylus brasiliensis: histochemical changes in the composition of mucins in goblet cells during infection in rats. Exp. Parasitol. 65, 84–90 (1988).
Yamauchi, J. et al. Altered expression of goblet cell- and mucin glycosylation-related genes in the intestinal epithelium during infection with the nematode Nippostrongylus brasiliensis in rat. APMIS 114, 270–278 (2006).
Oinuma, T., Abe, T., Nawa, Y., Kawano, J. & Suganuma, T. Glycoconjugates in rat small intestinal mucosa during infection with the intestinal nematode Nippostrongylus brasiliensis. Adv. Exp. Med. Biol. 371B, 975–978 (1995).
Holmen, J.M., Olson, F.J., Karlsson, H. & Hansson, G.C. Two glycosylation alterations of mouse intestinal mucins due to infection caused by the parasite Nippostrongylus brasiliensis. Glycoconj. J. 19, 67–75 (2002).
Karlsson, N.G. et al. Identification of transient glycosylation alterations of sialylated mucin oligosaccharides during infection by the rat intestinal parasite Nippostrongylus brasiliensis. Biochem. J. 350, 805–814 (2000).
Olson, F.J. et al. Blood group A glycosyltransferase occurring as alleles with high sequence difference is transiently induced during a Nippostrongylus brasiliensis parasite infection. J. Biol. Chem. 277, 15044–15052 (2002).
Khan, W.I., Abe, T., Ishikawa, N., Nawa, Y. & Yoshimura, K. Reduced amount of intestinal mucus by treatment with anti-CD4 antibody interferes with the spontaneous cure of Nippostrongylus brasiliensis-infection in mice. Parasite Immunol. 17, 485–491 (1995).
Kawai, Y. et al. T cell-dependent and -independent expression of intestinal epithelial cell-related molecules in rats infected with the nematode Nippostrongylus brasiliensis. APMIS 115, 210–217 (2007).
Salyers, A. & Whitt, D. Bacterial Pathogenesis: A Molecular Approach 2nd edn (ASM Press, Washington, DC, 1994 ).
Linden, S., Mahdavi, J., Hedenbro, J., Boren, T. & Carlstedt, I. Effects of pH on Helicobacter pylori binding to human gastric mucins: identification of binding to non-MUC5AC mucins. Biochem. J. 384, 263–270 (2004).
Teneberg, S. et al. Carbohydrate binding specificity of the neutrophil-activating protein of Helicobacter pylori. J. Biol. Chem. 272, 19067–19071 (1997).
Mahdavi, J. et al. Helicobacter pylori SabA adhesin in persistent infection and chronic inflammation. Science 297, 573–578 (2002).
Chen, C.C., Baylor, M. & Bass, D.M. Murine intestinal mucins inhibit rotavirus infection. Gastroenterology 105, 84–92 (1993).
Yolken, R.H., Ojeh, C., Khatri, I.A., Sajjan, U. & Forstner, J.F. Intestinal mucins inhibit rotavirus replication in an oligosaccharide-dependent manner. J. Infect. Dis. 169, 1002–1006 (1994).
Hytonen, J., Haataja, S. & Finne, J. Streptococcus pyogenes glycoprotein-binding strepadhesin activity is mediated by a surface-associated carbohydrate-degrading enzyme, Pullulanase. Infect. Immun. 71, 784–793 (2003).
Schwegmann, C., Zimmer, G., Yoshino, T., Enss, M. & Herrler, G. Comparison of the sialic acid binding activity of transmissible gastroenteritis coronavirus and E. coli K99. Virus Res. 75, 69–73 (2001).
Bisaillon, M., Senechal, S., Bernier, L. & Lemay, G. A glycosyl hydrolase activity of mammalian reovirus sigma1 protein can contribute to viral infection through a mucus layer. J. Mol. Biol. 286, 759–773 (1999).
Helander, A. et al. The viral sigma1 protein and glycoconjugates containing alpha2-3-linked sialic acid are involved in type 1 reovirus adherence to M cell apical surfaces. J. Virol. 77, 7964–7977 (2003).
Matrosovich, M. & Klenk, H.D. Natural and synthetic sialic acid-containing inhibitors of influenza virus receptor binding. Rev. Med. Virol. 13, 85–97 (2003).
Alexander, D.A. & Dimock, K. Sialic acid functions in enterovirus 70 binding and infection. J. Virol. 76, 11265–11272 (2002).
Willoughby, R.E. Rotaviruses preferentially bind O-linked sialylglycoconjugates and sialomucins. Glycobiology 3, 437–445 (1993).
Couceiro, J.N., Paulson, J.C. & Baum, L.G. Influenza virus strains selectively recognize sialyloligosaccharides on human respiratory epithelium; the role of the host cell in selection of hemagglutinin receptor specificity. Virus Res. 29, 155–165 (1993).
Babal, P. & Russell, L.C. Sialic acid-specific lectin-mediated adhesion of Tritrichomonas foetus and Tritrichomonas mobilensis. J. Parasitol. 85, 33–40 (1999).
Ryan, P.A., Pancholi, V. & Fischetti, V.A. Group A streptococci bind to mucin and human pharyngeal cells through sialic acid-containing receptors. Infect. Immun. 69, 7402–7412 (2001).
Velcich, A. et al. Colorectal cancer in mice genetically deficient in the mucin Muc2. Science 295, 1726–1729 (2002).
Van der Sluis, M. et al. Muc2-deficient mice spontaneously develop colitis, indicating that MUC2 is critical for colonic protection. Gastroenterology 131, 117–129 (2006).
Walters, R.W., Pilewski, J.M., Chiorini, J.A. & Zabner, J. Secreted and transmembrane mucins inhibit gene transfer with AAV4 more efficiently than AAV5. J. Biol. Chem. 277, 23709–23713 (2002).
Arcasoy, S.M. et al. MUC1 and other sialoglycoconjugates inhibit adenovirus mediated gene transfer to epithelial cells. Am. J. Respir. Cell Mol. Biol. 17, 422–435 (1997).
Peterson, J.A., Patton, S. & Hamosh, M. Glycoproteins of the human milk fat globule in the protection of the breast-fed infant against infections. Biol. Neonate 74, 143–162 (1998).
Yolken, R.H. et al. Human milk mucin inhibits rotavirus replication and prevents experimental gastroenteritis. J. Clin. Invest. 90, 1984–1991 (1992).
Dekker, J. & Tytgat, K.M. Binding of rotavirus by a 46 kD milk-glycoprotein may prevent gastroenteritis. J. Pediatr. Gastroenterol. Nutr. 17, 228–230 (1993).
DeSouza, M.M. et al. MUC1/episialin: a critical barrier in the female reproductive tract. J. Reprod. Immunol. 45, 127–158 (1999).
Kardon, R. et al. Bacterial conjunctivitis in Muc1 null mice. Invest. Ophthalmol. Vis. Sci. 40, 1328–1335 (1999).
Danjo, Y., Hazlett, L.D. & Gipson, I.K. C57BL/6 mice lacking Muc1 show no ocular surface phenotype. Invest. Ophthalmol. Vis. Sci. 41, 4080–4084 (2000).
McGuckin, M.A. et al. Muc1 mucin limits both Helicobacter pylori colonization of the murine gastric mucosa and associated gastritis. Gastroenterology 133, 1210–1218 (2007).
Vinall, L.E. et al. Altered expression and allelic association of the hypervariable membrane mucin MUC1 in Helicobacter pylori gastritis. Gastroenterology 123, 41–49 (2002).
Carvalho, F. et al. MUC1 gene polymorphism and gastric cancer—an epidemiological study. Glycoconj. J. 14, 107–111 (1997).
Silva, F. et al. MUC1 polymorphism confers increased risk for intestinal metaplasia in a Colombian population with chronic gastritis. Eur. J. Hum. Genet. 11, 380–384 (2003).
Blalock, T.D. et al. Functions of MUC16 in corneal epithelial cells. Invest. Ophthalmol. Vis. Sci. 48, 4509–4518 (2007).
Lu, W. et al. Cutting edge: enhanced pulmonary clearance of Pseudomonas aeruginosa by Muc1 knockout mice. J. Immunol. 176, 3890–3894 (2006).
Lillehoj, E.P. et al. Muc1 mucins on the cell surface are adhesion sites for Pseudomonas aeruginosa. Am. J. Physiol. Lung Cell Mol. Physiol. 280, L181–L187 (2001).
Lillehoj, E.P., Kim, B.T. & Kim, K.C. Identification of Pseudomonas aeruginosa flagellin as an adhesin for Muc1 mucin. Am. J. Physiol. Lung Cell Mol. Physiol. 282, L751–L756 (2002).
Kato, K., Lu, W., Kai, H. & Kim, K.C. Phosphoinositide 3-kinase is activated by MUC1 but not responsible for MUC1-induced suppression of TLR5 signaling. Am. J. Physiol. Lung Cell Mol. Physiol. 293, L686–L692 (2007).
Kawakubo, M. et al. Natural antibiotic function of a human gastric mucin against Helicobacter pylori infection. Science 305, 1003–1006 (2004).
Gururaja, T.L. et al. Candidacidal activity prompted by N-terminus histatin-like domain of human salivary mucin (MUC7). Biochim. Biophys. Acta Prot. Struct. Mol. Enzymol. 1431, 107–119 (1999).
Bobek, L.A. & Situ, H. MUC7 20-Mer: investigation of antimicrobial activity, secondary structure, and possible mechanism of antifungal action. Antimicrob. Agents Chemother. 47, 643–652 (2003).
Bruno, L.S. et al. Two-hybrid analysis of human salivary mucin MUC7 interactions. Biochim. Biophys. Acta 1746, 65–72 (2005).
Iontcheva, I., Oppenheim, F.G., Offner, G.D. & Troxler, R.F. Molecular mapping of statherin- and histatin-binding domains in human salivary mucin MG1 (MUC5B) by the yeast two-hybrid system. J. Dent. Res. 79, 732–739 (2000).
Royle, L. et al. Secretory IgA N- and O-glycans provide a link between the innate and adaptive immune systems. J. Biol. Chem. 278, 20140–20153 (2003).
Phalipon, A. et al. Secretory component: a new role in secretory IgA-mediated immune exclusion in vivo. Immunity 17, 107–115 (2002).
Salathe, M., Forteza, R. & Conner, G.E. Post-secretory fate of host defence components in mucus. Novartis Found Symp. 248, 20–26 (2002).
Forteza, R. et al. Hyaluronan serves a novel role in airway mucosal host defense. FASEB J. 15, 2179–2186 (2001).
Josenhans, C. & Suerbaum, S. The role of motility as a virulence factor in bacteria. Int. J. Med. Microbiol. 291, 605–614 (2002).
Moens, S. & Vanderleyden, J. Functions of bacterial flagella. Crit. Rev. Microbiol. 22, 67–100 (1996).
Ottemann, K.M. & Lowenthal, A.C. Helicobacter pylori uses motility for initial colonization and to attain robust infection. Infect. Immun. 70, 1984–1990 (2002).
Corfield, A.P., Wagner, S.A., Clamp, J.R., Kriaris, M.S. & Hoskins, L.C. Mucin degradation in the human colon: production of sialidase, sialate O-acetylesterase, N-acetylneuraminate lyase, arylesterase, and glycosulfatase activities by strains of fecal bacteria. Infect. Immun. 60, 3971–3978 (1992).
Haider, K. et al. Production of mucinase and neuraminidase and binding of Shigella to intestinal mucin. J. Diarrhoeal Dis. Res. 11, 88–92 (1993).
Corfield, A.P. et al. The roles of enteric bacterial sialidase, sialate O-acetyl esterase and glycosulfatase in the degradation of human colonic mucin. Glycoconj. J. 10, 72–81 (1993).
Homer, K.A., Whiley, R.A. & Beighton, D. Production of specific glycosidase activities by Streptococcus intermedius strain UNS35 grown in the presence of mucin. J. Med. Microbiol. 41, 184–190 (1994).
Roberton, A.M. & Wright, D.P. Bacterial glycosulphatases and sulphomucin degradation. Canad. J. Gastroenterol. 11, 361–366 (1997).
Lidell, M.E., Moncada, D.M., Chadee, K. & Hansson, G.C. Entamoeba histolytica cysteine proteases cleave the MUC2 mucin in its C-terminal domain and dissolve the protective colonic mucus gel. Proc. Natl. Acad. Sci. USA 103, 9298–9303 (2006).
Allen, A., Flemstrom, G., Garner, A. & Kivilaakso, E. Gastroduodenal mucosal protection. Physiol. Rev. 73, 823–857 (1993).
Silva, A.J., Pham, K. & Benitez, J.A. Haemagglutinin/protease expression and mucin gel penetration in El Tor biotype Vibrio cholerae. Microbiology 149, 1883–1891 (2003).
Solnick, J.V., Hansen, L.M., Salama, N.R., Boonjakuakul, J.K. & Syvanen, M. Modification of Helicobacter pylori outer membrane protein expression during experimental infection of rhesus macaques. Proc. Natl. Acad. Sci. USA 101, 2106–2111 (2004).
Siebers, A. & Finlay, B.B. M cells and the pathogenesis of mucosal and systemic infections. Trends Microbiol. 4, 22–29 (1996).
Neutra, M.R., Mantis, N.J., Frey, A. & Giannasca, P.J. The composition and function of M cell apical membranes: implications for microbial pathogenesis. Semin. Immunol. 11, 171–181 (1999).
Lelouard, H., Reggio, H., Mangeat, P., Neutra, M. & Montcourrier, P. Mucin-related epitopes distinguish M cells and enterocytes in rabbit appendix and Peyer's patches. Infect. Immun. 67, 357–367 (1999).
Lelouard, H. et al. Glycocalyx on rabbit intestinal M cells displays carbohydrate epitopes from Muc2. Infect. Immun. 69, 1061–1071 (2001).
Vazquez-Torres, A. & Fang, F.C. Cellular routes of invasion by enteropathogens. Curr. Opin. Microbiol. 3, 54–59 (2000).
Jones, B., Pascopella, L. & Falkow, S. Entry of microbes into the host: using M cells to break the mucosal barrier. Curr. Opin. Immunol. 7, 474–478 (1995).
Sakaguchi, T., Kohler, H., Gu, X., McCormick, B.A. & Reinecker, H.C. Shigella flexneri regulates tight junction-associated proteins in human intestinal epithelial cells. Cell Microbiol. 4, 367–381 (2002).
Goosney, D.L., Gruenheid, S. & Finlay, B.B. Gut feelings: enteropathogenic E. coli (EPEC) interactions with the host. Annu. Rev. Cell Dev. Biol. 16, 173–189 (2000).
Katz, J., Sambandam, V., Wu, J.H., Michalek, S.M. & Balkovetz, D.F. Characterization of Porphyromonas gingivalis-induced degradation of epithelial cell junctional complexes. Infect. Immun. 68, 1441–1449 (2000).
Amieva, M.R. et al. Disruption of the epithelial apical-junctional complex by Helicobacter pylori CagA. Science 300, 1430–1434 (2003).
Spicer, A.P., Rowse, G.J., Lidner, T.K. & Gendler, S.J. Delayed mammary tumor progression in Muc-1 null mice. J. Biol. Chem. 270, 30093–30101 (1995).
Heazlewood, C.K. et al. Aberrant mucin assembly in mice causes ER stress and spontaneous inflammation resembling ulcerative colitis. PLoS Med. 5, e54 (2008).
Sjunnesson, H., Sturegard, E., Grubb, A., Willen, R. & Wadstrom, T. Comparative study of Helicobacter pylori infection in guinea pigs and mice—elevation of acute-phase protein C3 in infected guinea pigs. FEMS Immunol. Med. Microbiol. 30, 167–172 (2001).
Kim, D.H. et al. Long-term evaluation of mice model infected with Helicobacter pylori: focus on gastric pathology including gastric cancer. Aliment Pharmacol. Ther. 18 (Suppl 1), 14–23 (2003).
Sturegard, E. et al. Severe gastritis in guinea-pigs infected with Helicobacter pylori. J. Med. Microbiol. 47, 1123–1129 (1998).
Dubois, A. et al. Transient and persistent experimental infection of nonhuman primates with Helicobacter pylori: implications for human disease. Infect. Immun. 64, 2885–2891 (1996).
Dubois, A. et al. Host specificity of Helicobacter pylori strains and host responses in experimentally challenged nonhuman primates. Gastroenterology 116, 90–96 (1999).
Dubois, A. et al. Seroepizootiology of Helicobacter pylori gastric infection in nonhuman primates housed in social environments. J. Clin. Microbiol. 33, 1492–1495 (1995).
Dubois, A. et al. Natural gastric infection with Helicobacter pylori in monkeys: a model for spiral bacteria infection in humans. Gastroenterology 106, 1405–1417 (1994).
Linden, S.K., Driessen, K.M. & McGuckin, M.A. Improved in vitro model systems for gastrointestinal infection by choice of cell line, pH, microaerobic conditions and optimization of culture conditions. Helicobacter 12, 341–353 (2007).
Cottet, S., Corthesy-Theulaz, I., Spertini, F. & Corthesy, B. Microaerophilic conditions permit to mimic in vitro events occurring during in vivo Helicobacter pylori infection and to identify Rho/Ras-associated proteins in cellular signaling. J. Biol. Chem. 277, 33978–33986 (2002).
Thornton, D.J. et al. Characterization of mucins from cultured normal human tracheobronchial epithelial cells. Am. J. Physiol. Lung Cell Mol. Physiol. 278, L1118–L1128 (2000).
Smoot, D.T. et al. Development of a human stomach explant organ culture system to study the pathogenesis of Helicobacter pylori. Digestion 46, 46–54 (1990).
Olfat, F.O., Naslund, E., Freedman, J., Boren, T. & Engstrand, L. Cultured human gastric explants: a model for studies of bacteria-host interaction during conditions of experimental Helicobacter pylori infection. J. Infect. Dis. 186, 423–427 (2002).
Haque, A. et al. Early interactions of Salmonella enterica serovar with human small intestinal epithelial explants. Gut 53, 1424–1430 (2004).
Gum, J.R. et al. Molecular cloning of human intestinal mucin cDNAs. Sequence analysis and evidence for genetic polymorphisms. J. Biol. Chem. 264, 6480–6487 (1989).
Ogata, S., Uehara, H., Chen, A. & Itzkowitz, S.H. Mucin gene expression in colonic tissues and cell lines. Cancer Res. 52, 5971–5978 (1992).
Dohrman, A. et al. Distribution of lysozyme and mucin (MUC2 and MUC3) mRNA in human bronchus. Exp. Lung Res. 20, 367–380 (1994).
Berry, M., Ellingham, R.B. & Corfield, A.P. Human preocular mucins reflect changes in surface physiology. Br. J. Ophthalmol. 88, 377–383 (2004).
Kerschner, J.E. Mucin gene expression in human middle ear epithelium. Laryngoscope 117, 1666–1676 (2007).
Porchet, N. et al. Human mucin genes: genomic organization and expression of MUC4, MUC5AC and MUC5B. Biochem. Soc. Trans. 23, 800–805 (1995).
Inatomi, T. et al. Expression of secretory mucin genes by human conjunctival epithelia. Invest. Ophthalmol. Vis. Sci. 37, 1684–1692 (1996).
Gipson, I.K. et al. Mucin genes expressed by human female reproductive tract epithelia. Biol. Reprod. 56, 999–1011 (1997).
Spurr-Michaud, S., Argueso, P. & Gipson, I. Assay of mucins in human tear fluid. Exp. Eye Res. 84, 939–950 (2007).
Gipson, I.K. et al. The Amount of MUC5B mucin in cervical mucus peaks at midcycle. J. Clin. Endocrinol. Metab. 86, 594–600 (2001).
Escande, F., Porchet, N., Aubert, J.P. & Buisine, M.P. The mouse Muc5b mucin gene: cDNA and genomic structures, chromosomal localization and expression. Biochem. J. 363, 589–598 (2002).
Davies, J.R. et al. Respiratory tract mucins: structure and expression patterns. Novartis Found Symp. 248, 76–88 (2002).
Vandenhaute, B. et al. Mucin gene expression in biliary epithelial cells. J. Hepatol. 27, 1057–1066 (1997).
Russo, C.L. et al. Mucin gene expression in human male urogenital tract epithelia. Hum. Reprod. 21, 2783–2793 (2006).
Toribara, N.W. et al. Human gastric mucin. Identification of a unique species by expression cloning. J. Biol. Chem. 268, 5879–5885 (1993).
Debolos, C., Garrido, M. & Real, F.X. MUC6 apomucin shows a distinct normal tissue distribution that correlates with lewis antigen expression in the human stomach. Gastroenterology 109, 723–734 (1995).
Bartman, A.E. et al. The MUC6 secretory mucin gene is expressed in a wide variety of epithelial tissues. J. Pathol. 186, 398–405 (1998).
Yu, D.F. et al. MUC19 expression in human ocular surface and lacrimal gland and its alteration in Sjogren syndrome patients. Exp. Eye Res. 86, 403–411 (2007).
Bobek, L.A., Tsai, H., Biesbrock, A.R. & Levine, M.J. Molecular cloning, sequence, and specificity of expression of the gene encoding the low molecular weight human salivary mucin (MUC7). J. Biol. Chem. 268, 20563–20569 (1993).
Sharma, P. et al. MUC5B and MUC7 are differentially expressed in mucous and serous cells of submucosal glands in human bronchial airways. Am. J. Respir. Cell Mol. Biol. 19, 30–37 (1998).
Pinkus, G.S. & Kurtin, P.J. Epithelial membrane antigen—a diagnostic discriminant in surgical pathology: immunohistochemical profile in epithelial, mesenchymal, and hematopoietic neoplasms using paraffin sections and monoclonal antibodies. Hum. Pathol. 16, 929–940 (1985).
Swallow, D.M. et al. The human tumour-associated epithelial mucins are coded by an expressed hypervariable gene locus PUM. Nature 328, 82–84 (1987).
Wreschner, D.H. et al. Human epithelial tumor antigen cDNA sequences. Differential splicing may generate multiple protein forms. Eur. J. Biochem. 189, 463–473 (1990).
Ho, S.B. et al. Mucin gene expression in normal, preneoplastic, and neoplastic human gastric epithelium. Cancer Res. 55, 2681–2690 (1995).
Gipson, I.K. & Inatomi, T. Mucin genes expressed by the ocular surface epithelium. Prog. Retinal Eye Res. 16, 81–98 (1997).
Wykes, M. et al. MUC1 epithelial mucin (CD227) is expressed by activated dendritic cells. J. Leukoc. Biol. 72, 692–701 (2002).
Ho, S.B. et al. Heterogeneity of mucin gene expression in normal and neoplastic tissues. Cancer Res. 53, 641–651 (1993).
Pratt, W.S. et al. Multiple transcripts of MUC3: evidence for two genes, MUC3A and MUC3B. Biochem. Biophys. Res. Commun. 275, 916–923 (2000).
Porchet, N. et al. Molecular cloning and chromosomal localization of a novel human tracheo-bronchial mucin cDNA containing tandemly repeated sequences of 48 base pairs. Biochem. Biophys. Res. Commun. 175, 414–422 (1991).
Moniaux, N. et al. Alternative splicing generates a family of putative secreted and membrane-associated MUC4 mucins. Eur. J. Biochem. 267, 4536–4544 (2000).
Gipson, I.K. et al. MUC4 and MUC5b transcripts are the prevalent mucin messenger ribonucleic acids of the human endocervix. Biol. Reprod. 60, 58–64 (1999).
Packer, L.M., Williams, S.J., Callaghan, S., Gotley, D.C. & McGuckin, M.A. Expression of the cell surface mucin gene family in adenocarcinomas. Int. J. Oncol. 25, 1119–1126 (2004).
Pallesen, L.T., Berglund, L., Rasmussen, L.K., Petersen, T.E. & Rasmussen, J.T. Isolation and characterization of MUC15, a novel cell membrane-associated mucin. Eur. J. Biochem. 269, 2755–2763 (2002).
Yin, B.W. & Lloyd, K.O. Molecular cloning of the CA125 ovarian cancer antigen: identification as a new mucin, MUC16. J. Biol. Chem. 276, 27371–27375 (2001).
Hori, Y., Spurr-Michaud, S., Russo, C.L., Argueso, P. & Gipson, I.K. Differential regulation of membrane-associated mucins in the human ocular surface epithelium. Invest. Ophthalmol. Vis. Sci. 45, 114–122 (2004).
Andersch-Bjorkman, Y., Thomsson, K.A., Holmen Larsson, J.M., Ekerhovd, E. & Hansson, G.C. Large scale identification of proteins, mucins, and their O-glycosylation in the endocervical mucus during the menstrual cycle. Mol. Cell Proteomics 6, 708–716 (2007).
Gipson, I.K. et al. MUC16 is lost from the uterodome (pinopode) surface of the receptive human endometrium: in vitro evidence that MUC16 is a barrier to trophoblast adherence. Biol. Reprod. 78, 134–142 (2008).
Gum, J.R. Jr, Crawley, S.C., Hicks, J.W., Szymkowski, D.E. & Kim, Y.S. MUC17, a novel membrane-tethered mucin. Biochem. Biophys. Res. Commun. 291, 466–475 (2002).
Higuchi, T. et al. Molecular cloning, genomic structure, and expression analysis of MUC20, a novel mucin protein, up-regulated in injured kidney. J. Biol. Chem. 279, 1968–1979 (2004).
Kubiet, M., Ramphal, R., Weber, A. & Smith, A. Pilus-mediated adherence of Haemophilus influenzae to human respiratory mucins. Infect. Immun. 68, 3362–3367 (2000).
Scharfman, A. et al. Adhesion of Pseudomonas aeruginosa to respiratory mucins and expression of mucin-binding proteins are increased by limiting iron during growth. Infect. Immun. 64, 5417–5420 (1996).
Shuter, J., Hatcher, V.B. & Lowy, F.D. Staphylococcus aureus binding to human nasal mucin. Infect. Immun. 64, 310–318 (1996).
Reddy, M.S. Binding of the pili of Pseudomonas aeruginosa to a low-molecular-weight mucin and neutral cystatin of human submandibular-sublingual saliva. Curr. Microbiol. 37, 395–402 (1998).
Hoffman, M.P. & Haidaris, C.G. Analysis of Candida albicans adhesion to salivary mucin. Infect. Immun. 61, 1940–1949 (1993).
Veerman, E.C. et al. Sulfated glycans on oral mucin as receptors for Helicobacter pylori. Glycobiology 7, 737–743 (1997).
Murray, P.A., Prakobphol, A., Lee, T., Hoover, C.I. & Fisher, S.J. Adherence of oral streptococci to salivary glycoproteins. Infect. Immun. 60, 31–38 (1992).
Prakobphol, A. et al. Separate oligosaccharide determinants mediate interactions of the low-molecular-weight salivary mucin with neutrophils and bacteria. Biochem. 38, 6817–6825 (1999).
Liu, B. et al. The recombinant N-terminal region of human salivary mucin MG2 (MUC7) contains a binding domain for oral Streptococci and exhibits candidacidal activity. Biochem. J. 345, 557–564 (2000).
Liu, B. et al. Interaction of human salivary mucin MG2, its recombinant N-terminal region and a synthetic peptide with Actinobacillus actinomycetemcomitans. J. Periodontal. Res. 37, 416–424 (2002).
Bosch, J.A. et al. Stress as a determinant of saliva-mediated adherence and coadherence of oral and nonoral microorganisms. Psychosom. Med. 65, 604–612 (2003).
Hirmo, S., Artursson, E., Puu, G., Wadstrom, T. & Nilsson, B. Helicobacter pylori interactions with human gastric mucin studied with a resonant mirror biosensor. J. Microbiol. Methods 37, 177–182 (1999).
Linden, S. et al. Strain- and blood group-dependent binding of Helicobacter pylori to human gastric MUC5AC glycoforms. Gastroenterology 123, 1923–1930 (2002).
Sun, R., Anderson, T.J., Erickson, A.K., Nelson, E.A. & Francis, D.H. Inhibition of adhesion of Escherichia coli k88ac fimbria to its receptor, intestinal mucin-type glycoproteins, by a monoclonal antibody directed against a variable domain of the fimbria. Infect. Immun. 68, 3509–3515 (2000).
Mack, D.R. & Blainnelson, P.L. Disparate in vitro inhibition of adhesion of enteropathogenic Escherichia coli rdec-1 by mucins isolated from various regions of the intestinal tract. Pediatr. Res. 37, 75–80 (1995).
Rajkumar, R., Devaraj, H. & Niranjali, S. Binding of Shigella to rat and human intestinal mucin. Mol. Cell Biochem. 178, 261–268 (1998).
Vimal, D.B., Khullar, M., Gupta, S. & Ganguly, N.K. Intestinal mucins: the binding sites for Salmonella typhimurium. Mol. Cell Biochem. 204, 107–117 (2000).
Jin, L.Z. & Zhao, X. Intestinal receptors for adhesive fimbriae of enterotoxigenic Escherichia coli (ETEC) K88 in swine—a review. Appl. Microbiol. Biotechnol. 54, 311–318 (2000).
Sylvester, F.A., Philpott, D., Gold, B., Lastovica, A. & Forstner, J.F. Adherence to lipids and intestinal mucin by a recently recognized human pathogen, Campylobacter upsaliensis. Infect. Immun. 64, 4060–4066 (1996).
Mantle, M. & Husar, S.D. Binding of Yersinia enterocolitica to purified, native small intestinal mucins from rabbits and humans involves interactions with the mucin carbohydrate moiety. Infect. Immun. 62, 1219–1227 (1994).
de Repentigny, L., Aumont, F., Bernard, K. & Belhumeur, P. Characterization of binding of Candida albicans to small intestinal mucin and its role in adherence to mucosal epithelial cells. Infect. Immun. 68, 3172–3179 (2000).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Linden, S., Sutton, P., Karlsson, N. et al. Mucins in the mucosal barrier to infection. Mucosal Immunol 1, 183–197 (2008). https://doi.org/10.1038/mi.2008.5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/mi.2008.5
This article is cited by
-
miR-125a-5p regulates the sialyltransferase ST3GAL1 in murine model of human intestinal campylobacteriosis
Gut Pathogens (2023)
-
Composition of mucus- and digesta-associated bacteria in growing pigs with and without diarrhea differed according to the presence of colonic inflammation
BMC Microbiology (2023)
-
ERdj5 protects goblet cells from endoplasmic reticulum stress-mediated apoptosis under inflammatory conditions
Experimental & Molecular Medicine (2023)
-
Basal stem cell progeny establish their apical surface in a junctional niche during turnover of an adult barrier epithelium
Nature Cell Biology (2023)
-
Functional roles of sphingolipids in immunity and their implication in disease
Experimental & Molecular Medicine (2023)