Abstract
Because of the small sizes of most viruses (typically 5–150 nm), standard optical microscopes, which have an optical diffraction limit of 200 nm, are not generally suitable for their direct observation. Electron microscopes usually require specimens to be placed under vacuum conditions, thus making them unsuitable for imaging live biological specimens in liquid environments. Indirect optical imaging of viruses has been made possible by the use of fluorescence optical microscopy that relies on the stimulated emission of light from the fluorescing specimens when they are excited with light of a specific wavelength, a process known as labeling or self-fluorescent emissions from certain organic materials. In this paper, we describe direct white-light optical imaging of 75-nm adenoviruses by submerged microsphere optical nanoscopy (SMON) without the use of fluorescent labeling or staining. The mechanism involved in the imaging is presented. Theoretical calculations of the imaging planes and the magnification factors have been verified by experimental results, with good agreement between theory and experiment.
Similar content being viewed by others
Introduction
Optical microscopic imaging resolution has a theoretical limit of approximately 200 nm within the visible light spectrum due to the far-field diffraction limit, which prevents the technique from being used for direct observation of live viruses (typically 5–150 nm, with some up to 300 nm). Progress in medical science and treatment of disease would benefit significantly from the availability of an instrument that enables direct optical imaging with a high resolution beyond the optical diffraction limit. Transmission electron microscopy (TEM) and scanning electron microscopy (SEM) are often used to image specially prepared dead virus structures at very high resolutions (<10 nm) in vacuum, but they are unsuitable for the study of live viruses or virus/cell interactions. Fluorescence optical microscopy is a recently established method for the imaging of cellular structures, bacteria and viruses beyond the optical diffraction limit, down to a resolution of 6 nm.1,2,3,4 This technique is based on the detection of light emitted by the fluorescing specimen when it is excited by light of a specific wavelength. Structured illumination, such as stimulated emission depletion and saturated structured illumination microscopy, which activate florescent light emission from a group of molecules simultaneously, are typically used.5,6 To enhance the imaging resolution, statistical reconstruction of the florescent light emission of single molecules at different times, such as statistical optical reconstruction microscopy and photo-activated localization microscopy have been developed,7,8 whereby an optically blurred image is sharpened through a deconvolution process, i.e., by digitally locating the peak of an optical profile or by using a point-spread function. The above fluorescent optical imaging techniques are confronted with the challenge of photobleaching, which limits the maximum time of light exposure to tens of seconds.9 In addition, fluorescent optical imaging techniques often require the conjugation of florescent molecules with the proteins of the target material, thus making such techniques somewhat intrusive, and only one type of stained protein can be imaged at a time, whereas there are over 10 000 types of proteins in each cell. Scanning near-field optical microscopy, which is based on point-by-point scanning of a nano-scale optical tip very close (within a few nanometers) to the target surface to illuminate the targets using the evanescent effect of the optical near-field, has demonstrated an imaging resolution of 60–100 nm.10 One of the drawbacks of the scanning near-field optical microscopy technique is the long time required to acquire the full image, thus making it difficult to study the dynamic behavior of viruses and cells. Imaging using a negative refractive index metamaterial has been shown to provide super-resolution capability down to 60 nm.11,12,13 The technique has, however, not yet been applied to cell/virus imaging. The high level of light attenuation is one of the key barriers to the practical application of superlens techniques for super-resolution biomedical imaging. A recent study using an X-ray femtosecond laser has shown super-resolution (subnanometer) imaging of virus particles just before their destruction.14 Super-oscillatory lens optical microscopy is another recently reported subwavelength imaging technique, and is based on a binary nano-structured mask.15 The imaging resolution so far is 105 nm (λ/6), and it is suitable for imaging an opaque target with transparent nano-structures. In 2011, a microsphere-coupled optical nanoscope was reported by some of the authors of this paper to have demonstrated an optical resolution of 50 nm using a SiO2 microsphere (with a diameter of 2–5 µm) in air for the imaging of inorganic materials.16 Until now, there has been no report demonstrating white light direct optical imaging of viruses below 100 nm in size.
In this paper, we report the use of submerged microsphere optical nanoscopy (SMON) for the direct imaging of an adenovirus with a diameter of 75 nm at a resolution beyond the optical diffraction limit. Large-diameter (100 µm) BaTiO3 spheres were used for this submerged optical imaging. The mechanisms involved in the dual-light large-microsphere submerged optical imaging are described.
Materials and methods
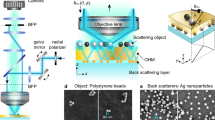
Imaging set-up
Optically transparent microspheres of BaTiO3 with diameters of 100 µm (supplied by Cospheric LLC; California, USA) were placed over the test specimen. Deionized water was deposited between the test specimen and the microscope lens. The optical microscope was used in both the reflection and transmission modes (Figure 1).
Adenovirus slide preparation
Replication-disabled adenoviruses of type 5 with deletions of the E1 and E3 genes were used. Adenovirus stock (109 MOI mL−1) was diluted 10 times in distilled H2O. For virus imaging using the BaTiO3 microsphere, the glass cover slide was first coated with a 5-nm-thick layer of gold to enable both optical and SEM imaging. One microliter of diluted adenovirus stock was then spread onto the gold-coated microscope slide and air-dried. The viruses were then fixed with one drop of 4% paraformaldehyde for 20 min, washed with tap water and then air-dried.
TEM imaging sample preparation
For TEM imaging, diluted adenoviruses were loaded onto a carbon-coated TEM grid, which was negative stained using uranyl acetate.
Results
Demonstration of super-resolution imaging by SMON
To demonstrate the super-resolution capability of large-microsphere (100 µm) SMON and to calibrate the imaging system, a Blu-Ray DVD disk with approximately 100-nm line spacing was imaged using an optical microscope (50×, NA=0.75) with a 100-µm diameter BaTiO3 microsphere in water under white light (reflective mode) illumination. The imaging result is shown in Figure 2, along with an image obtained by SEM for comparison. The structure (100-nm spacing) observed by the SMON technique is well beyond the spatial resolution limit of standard optical microscopes (approximately 200 nm).
To further demonstrate the resolution of the SMON imaging using large spheres submerged in water, an anodic aluminum oxide (AAO) sample was imaged with the 100-µm diameter BaTiO3 sphere coupled with a standard optical microscope submerged in water under the transmission light-illumination mode. The average pore size of the AAO sample was approximately 50 nm, as shown in the SEM image (Figure 3a). This structure was captured by the SMON imaging, as shown in Figure 3b, thus demonstrating the high-resolution imaging of the high refractive-index large-microsphere SMON approach with a 50-nm spatial resolution for images in water.
Virus imaging
TEM imaging was first used to confirm the size of the replication-disabled viruses, as shown in Figure 4, which indicates the size of an individual adenovirus to be approximately 75 nm.
Virus imaging by SMON was performed using 100-µm diameter BaTiO3 fused glass microspheres (with a refractive index of n=1.9) coupled to an optical microscope. An Olympus optical microscope with 80× and 100× objective lenses with NA=0.95 and NA=0.9 respectively in the reflection mode was used. The microsphere was placed on top of the viruses.
Figure 5a (SEM image) shows the place of interested with a cluster of viruses. The yellow arrow points a C shaped mark which was used to help with aligning the microsphere with viruses. The red arrow points to a shiny dust, which was helpful to find the right location of the viruses.
Images of adenovirus clusters. (a) An SEM image showing the place of interest. (b) An optical microscope image of a 100-µm BaTiO3 microsphere aligned on top of adenovirus clusters. (c) High magnification of SEM image of adenovirus clusters. (d) The low magnification SMON image of adenovirus clusters (100-µm BaTiO3 microsphere coupled with a 50× objective lens, NA=0.75). (e) A higher magnification of SEM image of adenovirus clusters. (f) A higher magnification SMON image of adenovirus clusters (100-µm BaTiO3 microsphere coupled with an 80× objective lens, NA=0.9). SEM, scanning electron microscopy; SMON, submerged microsphere optical nanoscopy.
Figure 5b confirms that a BaTiO3 microsphere was placed on top of viruses. This is a standard microscope image with a 50× objective lens. The imaging plane was set on the specimen surface.
By placing the microscopic imaging plane through the microsphere and into the specimen, a clearer SMON image (Figure 5d) could be obtained with a focusing distance of 40 µm below the top surface of the specimen. The red arrow indicates the location of a shiny dust in the SEM picture (Figure 5c). The viruses were, however, not seen with this optical set-up.
A higher-magnification SMON image was obtained by using a 80× objective lens with NA=0.9. The individual viruses were able to be resolved in Figure 5f. The imaging plane was set to 70 µm below the top surface of the specimen. Compared with the SEM image in Figure 5e, the SMON technique shows a better resolution for this organic sample. Non-conductive substances would cause electron charges during the SEM imaging which would directly affect its resolution.
Another higher magnification SMON image (Figure 6a) was obtained by using a 100-µm BaTiO3 microsphere coupled with an optical microscope with a 100× objective lens with NA=0.95. Figure 6b provides the quantitative intensity measurement from the reflective light emission. It clearly shows the peaks and separation between each spot.
(a) A SMON image of adenovirus clusters (100-µm BaTiO3 microsphere coupled with 100× objective lens, NA=0.95); the imaging plane was set 80 µm below the top surface of the specimen; (b) the quantitative contrast light intensity plot along the blue line indicated in SMON image. SMON, submerged microsphere optical nanoscopy.
Discussion
SMON imaging mechanisms
The Rayleigh criterion for the diffraction limit of a standard optical microscope states is given by the following:

where dmin is the diffraction-limited optical resolution, λ is the optical wavelength and NA is the numerical aperture of the microscope lens. For the optical microscopes used in this work, the theoretical diffraction limit resolutions of the light microscopes used in air for white light illumination (wavelengths in the range of 390–700 nm) would be in the range 250–449 nm for the NA=0.95 objective lens. In water (refractive index=1.33), these theoretical diffraction-limited imaging resolutions would be improved to 188–337 nm. Practical optical microscopes usually cannot reach the theoretical optical resolution due to spherical aberrations and imperfections in the optics. By using the microspheres, we have demonstrated optical resolution in the range of 50–120 nm in water. This resolution is clearly beyond the optical diffraction limits of these microscopes.
To understand the super-resolution effect, three-dimensional electromagnetic (EM) numerical (time-domain finite difference) simulations were performed using Computer Simulation Technology Microwave Studio software for two different sphere materials (BaTiO3 and polystyrene) and compared with the experimental data. An ideal diffraction limit model was developed, in which two 100-nm slits with a 50-nm pitch are embedded in a 20-nm thick perfect electric conductor in water. The materials of the microspheres used in the simulation are polystyrene (PS), with a refractive index of 1.59 (used in Figure 7a, 7b and 7d) and BaTiO3, with a refractive index of 1.9 (used in Figure 7e). The diameter of all the spheres is 10 µm. The substrate thickness is assumed to be 20 nm. The light wavelength is set to the center of the visible spectrum, i.e., 550 nm. The ambient condition in the model is water (refractive index n=1.33). When an EM plane wave (wavelength of 550 nm) arrives on the substrate, most of light energy is reflected, and only a small fraction is able to pass through the nano-slits (Figure 7a). The penetrated energy decays very rapidly and thus cannot be captured by a far-field detector. Based on the theory of Wolf and Nieto-Vesperinas,17 evanescent waves exist near the slits, and the fine structure information is encoded within this near field region.
Numerical simulation of microsphere-based nano-imaging in a water environment. (a) The scalar electric-field distribution when a parallel light beam is incident onto the substrate with sub-diffraction-limit fine structures, which represents the standard configuration of the microscope under the transmission mode. (b) The scalar electric-field distribution without considering the top semi-sphere reflection effect. (c) The schematic diagram of the top semi-sphere reflection in the transmission mode. (d) The scalar electric-field distribution considering the top semi-sphere reflection effect or by reflective mode light illumination, where the ‘reflected’ light or the illuminated light through the microsphere is simulated as two-sphere coupling. (e) The scalar electric-field distribution under the reflection mode.
When a PS microsphere is placed beneath the nano-slits in the water environment, a weak energy coupling caused by the conversion of the evanescent waves to propagating waves occurs, as shown in Figure 7b. This coupling occurs through the mechanism of frustrated total internal reflection.18,19 The converted propagating waves contain the high spatial-frequency information of the sub-wavelength slits.20 However, the converted propagating waves do not transmit beyond the sphere. This lack of transmission occurs when the imaging sample is under transmitted light-illumination mode. The super-resolution strength is mainly dependent on a narrow window of (n,q) parameters, where n is the refractive index of microsphere and q is the size parameter, defined as q=2πa/λ according to Mie theory.21 Figure 7c illustrates the light rays passing through the microsphere under the transmission imaging mode. When the transmitted light first passes into the microsphere, the light is focused as indicated by the black arrow lines. Some of the focused light is reflected at the interface of the top half of the sphere surface (as shown by the red arrow lines) and is refocused by the microsphere. During this process, the photonic nano-jets (i.e., a higher-intensity optical field located below the sphere that has a higher density spot beyond the optical diffraction limit) are generated, similar to the case in the reflection mode.16 The nano-jets arrive on the sample surface and illuminate the area below the microsphere at a high intensity and a high resolution, beyond the diffraction limit. This nano-jet super-resolution focusing phenomenon has been applied for laser nano-fabrication.22,23,24 By reversing this optical path, super-resolution imaging can be realized. The microsphere re-collects the light scattered by the virus/cells and converts the high spatial-frequency evanescent waves (no diffraction limits) into propagating waves that can be collected by far-field imaging. To illustrate this effect, a three-microsphere model is developed, as shown in Figure 7d, to decouple the projected and the received light through pure evanescent wave conversion. The top sphere is used to simulate the first pass of the light beam into the microsphere, i.e., the black arrow lines indicating the light rays, as shown in Figure 7c. The refocusing process of the reflected light from the top semi-sphere surface (i.e., the red arrow lines indicated in Figure 7c) or the reflected light projection from the optical microscope is simulated by the middle microsphere, as shown in Figure 7d. Photonic nano-jets are generated and redirected onto the substrate surface that is in contact with the bottom of the middle microsphere.
The microsphere converts the evanescent waves from the target surface into propagating waves. To clearly demonstrate this conversion process, a conjugate model was developed (Figure 7d) to decouple the scattered light from the incident light waves. The space beneath the substrate can be considered to be the conjugate ‘virtual’ space. The conjugate space has the same ‘structure’ as the real space shown in Figure 7c and is symmetric to the substrate surface. Therefore, the lowest microsphere in Figure 7d represents the conjugate space structure. The EM field in the conjugate space can be considered to be the collected EM field in the real space from the substrate surface without the background of the incident light waves. The substrate with two 100-nm slits and a 50-nm pitch is used as the fine structure. Based on this simulation set-up, the energy coupling from the evanescent waves into propagating waves can be clearly displayed in the transmission imaging mode or by using the reflection and dual reflection and transmission mode.
The above simulations indicate that the microsphere significantly enhances the energy penetration through subdiffraction-limit slits. This enhanced transmission can be attributed to the fact that the microspheres in the conjugate space (beneath the substrate) and in the real space (above the substrate) are close to each other. Hence, a phenomenon known as frustrated total internal reflection occurs, and the evanescent waves containing the high spatial-frequency information of the slits are linearly converted into propagating waves.18 Meanwhile, the photonic nano-jets generated by the two microspheres in the real space further enhance the energy coupling into the conjugate space; thus, the converted propagating waves have sufficient energy away from the microsphere for far-field imaging (as shown in Figure 7d). Therefore, the microsphere plays a dual-role for nano-imaging under both the transmission and reflection microscope imaging modes: (i) to generate photonic nano-jets by the top sphere for super-resolution target illumination and (ii) to convert the high spatial-frequency evanescent waves into magnified propagating waves for far-field detection.
To further understand the mechanism of subdiffraction-limit imaging via BaTiO3 microspheres in water in the reflection microscope imaging mode, a numerical simulation is performed using the three-dimensional finite-difference time-domain method for a 10-µm diameter BaTiO3 sphere submerged in water, as shown in Figure 7e. Considering the reflection configuration used in the imaging, the incident light first passes through the sphere and is focused by the microsphere. During this process, the sphere generates ‘photonic nano-jets’ with super-resolution.19,21,22,23 When the focused incident light illuminates the target surface, the reflected light can be recollected by the microsphere in the near-field. Using the conjugate model, the incident and reflected light waves are decoupled. As shown in Figure 7e, the upper half of the model is in real space, which describes the behavior of the light wave incident onto the target surface via the microsphere, whereas the lower half is the conjugate space, of which the ‘structure’ is same as real space and symmetric to the substrate surface. The conjugate space is used to show the reflected light wave without the background of the incident light waves. The microsphere transmits the energy coupled from the real space to the conjugate space through the subdiffraction-limit slits. The evanescent waves that contain the high spatial-frequency information (i.e., fine structures) are converted into propagating waves. The nano-photonic jets generated via the upper microsphere provide enough energy coupling to enable far-field imaging. Hence, subdiffraction-limit nano-imaging can be achieved using BaTiO3 microspheres in water for reflection mode microscopy. The simulation indicates that super-resolution imaging is possible in water with the aid of microspheres in both transmission and reflection modes.
The super-resolution virus imaging is obviously attributed to the existence of the transparent microspheres. We compare the SMON with super-solid immersion lens imaging to identify the differences. Based on the solid immersion lens theory, the maximum imaging resolution improvement by a microsphere should be n0/n1.25 The maximum imaging resolutions achieved using a solid immersion lens are 163 nm and 137 nm for the PS and BaTiO3 microspheres in water, respectively, for the microscope used in this study. Based on the super solid immersion lens effect, the magnification via the microsphere can be calculated as (n1/n0)2. For the PS microspheres (n1=1.60) in water (n1=1.33), the magnification should be 1.45. For the BaTiO3 microspheres (n1=1.9) in water, the magnification is approximately 2.0. The experimental values of the image magnifications in water by the microspheres are higher (1.5–3 for the PS spheres and 2.5–4 for the BaTiO3 spheres) than those by the solid immersion lens effect. The experimental imaging resolution is also higher than the theoretical limit of the solid immersion lens. Therefore, the microsphere plays an important role in transforming the high-resolution evanescent waves in the near field into far-field propagating waves.
Because there is no diffraction limit for evanescent waves, the only limitation is the means of transforming the near-field evanescent waves into far-field propagating waves with sufficient image magnification so that it falls within the optical resolution of the standard optical microscope used to collect the image. Therefore, image magnification is very important. In addition, the SMON imaging plane was found to not be on the target surface but well below it; i.e., the images are virtual.16 The imaging plane position and additional image magnification factor introduced by the microsphere can be determined by considering the spherical lens effect and spherical aberration, as follows:



where f is the focal length of the microsphere from the sphere center, d is the transverse distance from the optical axis, R is the microsphere radius, n0 is the ambient refractive index, n1 is the refractive index of the microsphere, s is the virtual imaging plane position from the center of the microsphere, M is the microsphere image magnification factor and δ is the distance from the target to the microsphere surface.
Table 1 lists the range of the paraxial focal length of various microspheres in air and water, where the virtual imaging plane position range caused by the spherical aberration was also considered. All the imaging planes are below the microsphere, and the microspheres in water have closer imaging planes to the sphere. The short imaging plane range could be beneficial to obtaining a clearer nano-image without significant spherical aberration blurring. Note that the paraxial imaging plane generated by the BaTiO3 microspheres is far away from the target surface in air and hence it cannot be captured by the objective lens used in this work. When the BaTiO3 was submerged into water, the paraxial imagine plane is located closer to the microsphere, with a distance that the objective lens can image.
Figure 8 illustrates the paraxial focal length, determined by Equation (2), and the imaging plane position, determined by Equaton (3).
Applying Equations (3) and (4), the magnification factors and the virtual imaging plane positions for the various microspheres used in this work can be determined. Tables 2 and 3 list the calculated results in comparison with the experimental values, where the effect of δ was considered in the magnification factor calculations. The theoretical results exhibit good agreement with the experimental results.
Engineering and practical aspects of SMON imaging of biomedical specimens
Because the microspheres have a curved surface close to the target, the distance between the microsphere and the target differs at different locations away from the center. It is important to know whether evanescent waves still exist towards the edges, where the distance from the sphere surface to the target surface is large. For example, when a 50-µm diameter microsphere is placed on a flat surface, the distance, x, separating the microsphere surface from the sample surface is increased to up to 700 nm when the lateral position is 6 µm from the central axis, as shown in Figure 9.
Considering the evanescent wave decay from the object surface, the wave vector kz of evanescent waves can be expressed as follows:

Substituting the evanescent form of the wave vector k, the transmitted wave can be written as follows:

When the electric field E≥E0/e, the evanescent wave energy can be coupled into the microsphere and converted into propagating waves. Therefore, the following equation must be satisfied to identify the maximum plane wave vector ( ) that can be collected by microspheres:
) that can be collected by microspheres:

where d is the distance between the object surface and the microsphere surface. From Equation (7), even if there is a distance between the object and the microsphere, some of the evanescent waves that contain high spatial-frequencies beyond diffraction limit can still be collected by the microsphere. However, the subdiffraction-limit frequencies that can be collected by the microsphere are dramatically reduced as the distance d increases. For example, when the distance is the light wavelength, the maximum plane wave vector ( ) is 1.01nω/c, i.e., the maximum resolution is improved by approximately 1.01 times. Therefore, the distance between object and microsphere surfaces should be on the same order as the light wavelength to obtain a significant imaging resolution improvement. In our experiment, the distance at a position 6 µm away from the 50-µm sphere center is calculated to be ∼700 nm, which is within the optical wavelength range of 400–700 nm. The SMON super-resolution can therefore be achieved for an imaged area within a radius of 6 µm.
) is 1.01nω/c, i.e., the maximum resolution is improved by approximately 1.01 times. Therefore, the distance between object and microsphere surfaces should be on the same order as the light wavelength to obtain a significant imaging resolution improvement. In our experiment, the distance at a position 6 µm away from the 50-µm sphere center is calculated to be ∼700 nm, which is within the optical wavelength range of 400–700 nm. The SMON super-resolution can therefore be achieved for an imaged area within a radius of 6 µm.
The use of a gold coating on the glass substrate was found to increase the imaging magnification and sharpness when a nano-structure existed, as in the AAO sample, because the surface plasmonic effect can enhance the evanescent waves.
The placement of the microsphere on the target surface ensures consistency in the distance between the sphere and the target. This is, however, not necessarily the optimum condition. In practical applications of the SMON, the microsphere needs to be attached to the optical microscope objective lens with a specific distance between the sphere and the objective lens, depending on the lens optical properties (e.g., focal length and numerical aperture) and the imaging media (water, oil or biological liquid). This distance also allows a small distance to be created between the sphere and the target distance (<1× optical wavelength) so that the target can move freely below the sphere. This paper demonstrates the principles and feasibility of the super-resolution imaging of biological samples. Further work is required to create a practical imaging device based on the SMON principle with the optimized microspheres attached to the microscope objective lens.
The microsphere diameter needs to be chosen carefully. As listed in Table 1, the paraxial imaging plane position depends on the microsphere diameter. A large microsphere can form the paraxial-imaging plane slightly away from the target surface. The resulting small separation of the imaging plane from the target surface reduces the blurring effect caused by the scattered light from the target surface.
The presence of liquid (especially the immersion of the objective lens) is important to allow effective coupling of the optical images from the microsphere to the optical microscope. For a BaTiO3 microsphere, no images could be obtained in air because the first virtual imaging plane was far away from the target surface (approximately 18R). Water can reduce the distance between the imaging plane and the surface; however, the imaging magnification factor is reduced compared with the dry condition due to the increased refractive index of the medium.
For biomedical imaging, there are several techniques available, primarily dominated by the use of fluorescent light optical microscopy, to achieve super-resolution. However, fluorescent light optical microscopy only images certain parts of the protein that emit fluorescent light. Holographic imaging is another technique that has been reported to be able to image biological materials, such as bacteria.24,26 Holographic imaging does not directly observe objects. Instead, it relies on the detection and digital processing of diffraction patterns when the object is illuminated. Based on the differences of the surface texture, analysis of the detected holograms provides information on the identities of different cell types. Therefore, the current application of holographic imaging in cell biology has been limited to cell sorting and identification, i.e., cytometry systems. Holographic imaging is able to distinguish bacteria from red blood cells in whole blood samples, which could be used for the diagnosis of infectious diseases.26 However, holographic imaging has not been able to acquire super-resolution images of cellular structures without being combined with an electron microscope or an atomic force microscope. Digital or computational holography has the advantage of acquiring high-contrast three-dimensional pictures of un-labeled cells.27 The use of two-dimensional evanescent standing wave illumination has been shown by Chung et al.27 to improve the image resolution of a standard total internal reflection fluorescent optical microscope to approximately 100 nm for cell imaging. A continuous-wave 532-nm laser was used for the illumination. Standard total internal reflection fluorescent microscopy has been demonstrated to be able to image sindbis virus and HIV virus. Total internal reflection fluorescent microscopy requires fluorescent staining. The use of white light illumination in the SMON imaging method enables the use of standard low-cost optical microscopes for super-resolution imaging if used in conjunction with suitable microspheres and the imaging procedures reported in this paper.
Conclusions
A simple method for direct white-light optical observation of 75-nm adenoviruses was demonstrated for the first time by coupling a standard optical microscope with a 100-µm diameter BaTiO3 microsphere in water without fluorescent particle labeling. The mechanism of super-resolution imaging using SMON was found to be based on the conversion of evanescent waves in the near field into the magnified prorogating waves in the far field by the microsphere through the mechanism of frustrated total internal reflection. Nano-jet formation through focusing of light by the microsphere plays an important role in enhancing the image contrast by delivering the converted propagating wave to the space outside the sphere. Because evanescent waves do not have a diffraction limit, the transformation of the evanescent wave to a magnified propagating wave within the optical resolution limit of a standard optical microscope is important. In other words, the resolution of the microsphere-based optical imaging, although theoretically unlimited, is limited in practice by the magnification factor of the microsphere in converting the evanescent wave to the propagating wave. Qualitative relationships to determine the imaging plane location (normally well below the microspheres) and the microsphere-induced magnification factors for the SMON were derived and compared to the experiments, with close agreement. This work opens new opportunities for the study of virus/cell/bacteria/drug interactions to better understand the causes of various deceases; super-resolution direct optical SMON imaging has the potential to become an alternative and complimentary technique to fluorescent optical microscopy and electron microscopy.
Author contributions
LL conceived the concept, led the research, participated in the virus imaging experiments and prepared the manuscript. TW prepared all the biological imaging samples, performed the TEM imaging of the viruses, participated in the optical imaging experiments and contributed to the manuscript preparation. WG performed the virus imaging experiment and contributed to the manuscript writing. YY performed the theoretical simulation work and contributed to the manuscript preparation, particularly on the Discussion section. SL performed the Blu-Ray and AAO imaging experiments, participated in the SMON biological sample imaging experiments and contributed to the manuscript preparation.
References
Hell SW . Far-field optical nanoscopy. Science 2007; 316: 1153–1158.
Huang B, Wang W, Bates M, Zhuang X . Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science 2008; 319: 810–813.
Stone MD, Mihalusova M, O'Connor CM, Prathapam R, Collins K et al. Stepwise protein-mediated RNA folding directs assembly of telomerase ribonucleoprotein. Nature 2007; 446: 458–461.
Huang B, Babcock H, Zhuang X . Breaking the diffraction barrier: super-resolution imaging of cells. Cell 2010; 143: 1047–1058.
Hell SW, Wichmann J . Breaking the diffraction resolution limit by stimulated emission: stimulated-emission-depletion fluorescence microscopy. Opt Lett 1994; 19: 780–782.
Heintzmann R, Jovin TM, Cremer C . Saturated patterned excitation microscopy—a concept for optical resolution improvement. J Opt Soc Am A Opt Image Sci Vis 2002; 19: 1599–1609.
Rust MJ, Bates M, Zhuang X . Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat Methods 2006; 3: 793–795.
Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science 2006; 313: 1642–1645.
Renn A, Seelig J, Sandoghdar V . Oxygen-dependent photochemistry of fluorescent dyes studied at the single molecule level. Mol Phys 2006; 104: 409–414.
Dürig U, Pohl DW, Rohner F . Near-field optical-scanning microscopy. J Appl Phys 1986; 59: 3318–3327.
Pendry JB . Negative refraction makes a perfect lens. Phys Rev Lett 2000; 85: 3966–3969.
Fang N, Lee H, Sun C, Zhang X . Sub-diffraction-limited optical imaging with a silver superlens. Science 2005; 308: 534–537.
Smolyaninov II, Davis CC, Elliott J, Wurtz GA, Zayats AV . Super-resolution optical microscopy based on photonic crystal materials. Phys Rev B 2005; 72: 085442.
Chapman HN, Fromme P, Barty A, White TA, Kirian RA et al. Femtosecond X-ray protein nanocrystallography. Nature 2011; 470: 73–78.
Roger ET, Lindberg J, Roy T, Savo S, Chad JE et al. A super-oscillatory lens optical microscope for subwavelength imaging. Nat Mater 2012; 11: 432–435.
Wang Z, Guo W, Li L, Luk'yanchuk B, Khan A et al. Optical virtual imaging at 50 nm lateral resolution with a white-light nanoscope. Nat Commun 2011; 2: 218.
Richards B, Wolf E . Electromagnetic diffraction in optical systems. II. Structure of the image field in an aplanatic system. Proc R Soc Lond A 1959; 253: 358–379.
Courjon D, Bainier C . Near field microscopy and near field optics. Rep Prog Phys 1994; 57: 989–1028.
Li L, Guo W, Wang ZB, Liu Z, Whitehead D et al. Large area laser nano-texturing with user-defined patterns. J Micromech Microeng 2009; 19: 064002.
Webb RH . Confocal optical microscopy. Rep Prog Phys 1996; 59: 427–471.
Wang Z, Joseph N, Li L, Luk'yanchuk BS . A review of optical near-fields in particle/tip-assisted laser nanofabrication. Proc IMechE Part C 2010; 224: 1113–1127.
Guo W, Wang ZB, Li L, Liu Z, Luk'yanchuk B . Chemical-assisted laser parallel nanostructuring of silicon in optical near fields. Nanotechnology 2008; 19: 455302.
Li L, Hong M, Schmidt M, Zhong M, Mashe M et al. Laser nano-manufacturing—state of the art and challenges (Keynote at 61st CIRP General Assembly, 21–27 August Budapest 2011). CIRP Ann 2011; 60: 735–755.
Seo S, Su TW, Tseng D, Erlinger A, Izcan A . Lens-free holographic imaging for on-chip cytometry and dianostics. Lab Chip 2009; 9: 777–787.
Serrels KA, Ramsay E, Dalgarno PA, Gerardot BD, O'Connor JA et al. Solid immersion lens applications for nanophotonic devices. J Nanophoton 2008; 2: 021854.
Moon I, Daneshpanah M, Anand A, Javidi B . Cell identification with computational 3D holographic microscopy. Opt Photon News 2011; 22: 18–23.
Chung E, Kim D, Cui Y, Kim YH, So PT . Two-dimensional standing wave total internal reflection fluorescence microscopy: superresolution imaging of single molecular and biological specimens. Biophys J 2007; 93: 1747–1757.
Acknowledgements
We thank Dr Peter March for his help in using microscopes in the Bio-imaging Facility, Dr Aleksandr Mironov for his help with the electron microscope Facility in the Faculty of Life Sciences, the University of Manchester, and Dr Zengbo Wang for technical discussions during the early stages of the work.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
This work is licensed under the Creative Commons Attribution-NonCommercial-No Derivative Works 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Li, L., Guo, W., Yan, Y. et al. Label-free super-resolution imaging of adenoviruses by submerged microsphere optical nanoscopy. Light Sci Appl 2, e104 (2013). https://doi.org/10.1038/lsa.2013.60
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/lsa.2013.60
Keywords
This article is cited by
-
Sub-50 nm optical imaging in ambient air with 10× objective lens enabled by hyper-hemi-microsphere
Light: Science & Applications (2023)
-
Wave optics of imaging with contact ball lenses
Scientific Reports (2023)
-
An acoustofluidic scanning nanoscope using enhanced image stacking and processing
Microsystems & Nanoengineering (2022)
-
Microspheres give improved resolution in nondestructive examination of semiconductor devices
Light: Science & Applications (2022)
-
Characteristic parameters of photonic nanojets of single dielectric microspheres illuminated by focused broadband radiation
Scientific Reports (2022)