Abstract
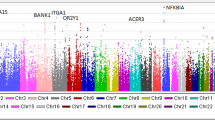
Immunoglobulin light chain (AL) amyloidosis is characterized by tissue deposition of amyloid fibers derived from immunoglobulin light chain. AL amyloidosis and multiple myeloma (MM) originate from monoclonal gammopathy of undetermined significance. We wanted to characterize germline susceptibility to AL amyloidosis using a genome-wide association study (GWAS) on 1229 AL amyloidosis patients from Germany, UK and Italy, and 7526 healthy local controls. For comparison with MM, recent GWAS data on 3790 cases were used. For AL amyloidosis, single nucleotide polymorphisms (SNPs) at 10 loci showed evidence of an association at P<10−5 with homogeneity of results from the 3 sample sets; some of these were previously documented to influence MM risk, including the SNP at the IRF4 binding site. In AL amyloidosis, rs9344 at the splice site of cyclin D1, promoting translocation (11;14), reached the highest significance, P=7.80 × 10−11; the SNP was only marginally significant in MM. SNP rs79419269 close to gene SMARCD3 involved in chromatin remodeling was also significant (P=5.2 × 10−8). These data provide evidence for common genetic susceptibility to AL amyloidosis and MM. Cyclin D1 is a more prominent driver in AL amyloidosis than in MM, but the links to aggregation of light chains need to be demonstrated.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Hemminki K, Li X, Forsti A, Sundquist J, Sundquist K . Incidence and survival in non-hereditary amyloidosis in Sweden. BMC Public Health 2012; 12: 974.
Pinney JH, Smith CJ, Taube JB, Lachmann HJ, Venner CP, Gibbs SD et al. Systemic amyloidosis in England: an epidemiological study. Br J Haematol 2013; 161: 525–532.
Blancas-Mejia LM, Ramirez-Alvarado M . Systemic amyloidoses. Annu Rev Biochem 2013; 82: 745–774.
Merlini G, Seldin DC, Gertz MA . Amyloidosis: pathogenesis and new therapeutic options. J Clin Oncol 2011; 29: 1924–1933.
Ramirez-Alvarado M . Amyloid formation in light chain amyloidosis. Curr Top Med Chem 2012; 12: 2523–2533.
Palladini G, Merlini G . What is new in diagnosis and management of light chain amyloidosis? Blood 2016; 128: 159–168.
Kyle RA, Durie BG, Rajkumar SV, Landgren O, Blade J, Merlini G et al. Monoclonal gammopathy of undetermined significance (MGUS) and smoldering (asymptomatic) multiple myeloma: IMWG consensus perspectives risk factors for progression and guidelines for monitoring and management. Leukemia 2010; 24: 1121–1127.
Merlini G, Palladini G . Differential diagnosis of monoclonal gammopathy of undetermined significance. Hematol Am Soc Hematol Educ Program 2012; 2012: 595–603.
Broderick P, Chubb D, Johnson DC, Weinhold N, Forsti A, Lloyd A et al. Common variation at 3p22.1 and 7p15.3 influences multiple myeloma risk. Nat Genet 2012; 44: 58–61.
Chubb D, Weinhold N, Broderick P, Chen B, Johnson DC, Forsti A et al. Common variation at 3q26.2, 6p21.33, 17p11.2 and 22q13.1 influences multiple myeloma risk. Nat Genet 2013; 45: 1221–1225.
Swaminathan B, Thorleifsson G, Joud M, Ali M, Johnsson E, Ajore R et al. Variants in ELL2 influencing immunoglobulin levels associate with multiple myeloma. Nat Commun 2015; 6: 7213.
Weinhold N, Forsti A, da Silva Filho MI, Nickel J, Campo C, Hoffmann P et al. Immunoglobulin light-chain amyloidosis shares genetic susceptibility with multiple myeloma. Leukemia 2014; 28: 2254–2256.
Weinhold N, Johnson DC, Chubb D, Chen B, Försti A, Hosking FJ et al. The CCND1 G870A polymorphism is a risk factor for t(11;14)(q13;q32) multiple myeloma. Nat Genet 2013; 45: 522–525.
Bochtler T, Hegenbart U, Cremer FW, Heiss C, Benner A, Hose D et al. Evaluation of the cytogenetic aberration pattern in amyloid light chain amyloidosis as compared with monoclonal gammopathy of undetermined significance reveals common pathways of karyotypic instability. Blood 2008; 111: 4700–4705.
Bochtler T, Hegenbart U, Heiss C, Benner A, Moos M, Seckinger A et al. Hyperdiploidy is less frequent in AL amyloidosis compared with monoclonal gammopathy of undetermined significance and inversely associated with translocation t(11;14). Blood 2011; 117: 3809–3815.
Harrison CJ, Mazzullo H, Ross FM, Cheung KL, Gerrard G, Harewood L et al. Translocations of 14q32 and deletions of 13q14 are common chromosomal abnormalities in systemic amyloidosis. Br J Haematol 2002; 117: 427–435.
Hayman SR, Bailey RJ, Jalal SM, Ahmann GJ, Dispenzieri A, Gertz MA et al. Translocations involving the immunoglobulin heavy-chain locus are possible early genetic events in patients with primary systemic amyloidosis. Blood 2001; 98: 2266–2268.
Mitchell JS, Li N, Weinhold N, Forsti A, Ali M, van Duin M et al. Genome-wide association study identifies multiple susceptibility loci for multiple myeloma. Nat Commun 2016; 7: 12050.
Schonland SO, Hegenbart U, Bochtler T, Mangatter A, Hansberg M, Ho AD et al. Immunohistochemistry in the classification of systemic forms of amyloidosis: a systematic investigation of 117 patients. Blood 2012; 119: 488–493.
Bochtler T, Hegenbart U, Kunz C, Benner A, Kimmich C, Seckinger A et al. Prognostic impact of cytogenetic aberrations in AL amyloidosis patients after high-dose melphalan: a long-term follow-up study. Blood 2016; 128: 594–602.
Kohler A, Chen B, Gemignani F, Elisei R, Romei C, Figlioli G et al. Genome-wide association study on differentiated thyroid cancer. J Clin Endocrinol Metab 2013; 98: E1674–E1681.
Howie BN, Donnelly P, Marchini J . A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet 2009; 5: e1000529.
Delaneau O, Marchini J, Zagury JF . A linear complexity phasing method for thousands of genomes. Nat Methods 2012; 9: 179–181.
Marchini J, Howie B, Myers S, McVean G, Donnelly P . A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet 2007; 39: 906–913.
Magi R, Morris AP . GWAMA: software for genome-wide association meta-analysis. BMC Bioinform 2010; 11: 288.
Bhattacharjee S, Rajaraman P, Jacobs KB, Wheeler WA, Melin BS, Hartge P et al. A subset-based approach improves power and interpretation for the combined analysis of genetic association studies of heterogeneous traits. Am J Hum Genet 2012; 90: 821–835.
Rosenbloom KR, Armstrong J, Barber GP, Casper J, Clawson H, Diekhans M et al. The UCSC Genome Browser database: 2015 update. Nucleic Acids Res 2015; 43: D670–D681.
Ward LD, Kellis M . HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res 2012; 40: D930–D934.
Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 2010; 26: 2336–2337.
Weinhold N, Meissner T, Johnson DC, Seckinger A, Moreaux J, Forsti A et al. The 7p15.3 (rs4487645) association for multiple myeloma shows strong allele-specific regulation of the MYC-interacting gene CDCA7L in malignant plasma cells. Haematologica 2015; 100: e110–e113.
Westra HJ, Peters MJ, Esko T, Yaghootkar H, Schurmann C, Kettunen J et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet 2013; 45: 1238–1243.
Knudsen KE, Diehl JA, Haiman CA, Knudsen ES . Cyclin D1: polymorphism, aberrant splicing and cancer risk. Oncogene 2006; 25: 1620–1628.
Millar EK, Dean JL, McNeil CM, O'Toole SA, Henshall SM, Tran T et al. Cyclin D1b protein expression in breast cancer is independent of cyclin D1a and associated with poor disease outcome. Oncogene 2009; 28: 1812–1820.
Olshavsky NA, Comstock CE, Schiewer MJ, Augello MA, Hyslop T, Sette C et al. Identification of ASF/SF2 as a critical, allele-specific effector of the cyclin D1b oncogene. Cancer Res 2011; 70: 3975–3984.
Fonseca R, Barlogie B, Bataille R, Bastard C, Bergsagel PL, Chesi M et al. Genetics and cytogenetics of multiple myeloma: a workshop report. Cancer Res 2004; 64: 1546–1558.
Palumbo A, Anderson K . Multiple myeloma. N Engl J Med 2011; 364: 1046–1060.
Pestell RG . New roles of cyclin D1. Am J Pathol 2013; 7// 183: 3–9.
Pabalan N, Bapat B, Sung L, Jarjanazi H, Francisco-Pabalan O, Ozcelik H . Cyclin D1 Pro241Pro (CCND1-G870A) polymorphism is associated with increased cancer risk in human populations: a meta-analysis. Cancer Epidemiol Biomarkers Prev 2008; 17: 2773–2781.
Musgrove EA, Caldon CE, Barraclough J, Stone A, Sutherland RL . Cyclin D as a therapeutic target in cancer. Nat Rev Cancer 2011; 11: 558–572.
De Silva NS, Simonetti G, Heise N, Klein U . The diverse roles of IRF4 in late germinal center B-cell differentiation. Immunol Rev 2012; 247: 73–92.
Pathak S, Ma S, Trinh L, Eudy J, Wagner KU, Joshi SS et al. IRF4 is a suppressor of c-Myc induced B cell leukemia. PLoS One 2011; 6: e22628.
Nutt SL, Hodgkin PD, Tarlinton DM, Corcoran LM . The generation of antibody-secreting plasma cells. Nat Rev Immunol 2015; 15: 160–171.
Tellier J, Shi W, Minnich M, Liao Y, Crawford S, Smyth GK et al. Blimp-1 controls plasma cell function through the regulation of immunoglobulin secretion and the unfolded protein response. Nat Immunol 2016; 17: 323–330.
Pallante P, Forzati F, Federico A, Arra C, Fusco A . Polycomb protein family member CBX7 plays a critical role in cancer progression. Am J Cancer Res 2015; 5: 1594–1601.
Jordan NV, Prat A, Abell AN, Zawistowski JS, Sciaky N, Karginova OA et al. SWI/SNF chromatin-remodeling factor Smarcd3/Baf60c controls epithelial-mesenchymal transition by inducing Wnt5a signaling. Mol Cell Biol 2013; 33: 3011–3025.
Lauc G, Huffman JE, Pucic M, Zgaga L, Adamczyk B, Muzinic A et al. Loci associated with N-glycosylation of human immunoglobulin G show pleiotropy with autoimmune diseases and haematological cancers. PLoS Genet 2013; 9: e1003225.
Menschikowski M, Hagelgans A, Schuler U, Froeschke S, Rosner A, Siegert G . Plasma levels of phospholipase A2-IIA in patients with different types of malignancies: prognosis and association with inflammatory and coagulation biomarkers. Pathol Oncol Res 2013; 19: 839–846.
Grudnik P, Bange G, Sinning I . Protein targeting by the signal recognition particle. Biol Chem 2009; 390: 775–782.
Karamyshev AL, Patrick AE, Karamysheva ZN, Griesemer DS, Hudson H, Tjon-Kon-Sang S et al. Inefficient SRP interaction with a nascent chain triggers a mRNA quality control pathway. Cell 2014; 156: 146–157.
Lee YJ, Kim HS, Ryu AJ, Jeong KJ . Enhanced production of full-length immunoglobulin G via the signal recognition particle (SRP)-dependent pathway in Escherichia coli. J Biotechnol 2013; 165: 102–108.
Auner HW, Cenci S . Recent advances and future directions in targeting the secretory apparatus in multiple myeloma. Br J Haematol 2015; 168: 14–25.
Xing Y, Igarashi H, Wang X, Sakaguchi N . Protein phosphatase subunit G5PR is needed for inhibition of B cell receptor-induced apoptosis. J Exp Med 2005; 202: 707–719.
Katayama K, Yamaguchi M, Noguchi K, Sugimoto Y . Protein phosphatase complex PP5/PPP2R3C dephosphorylates P-glycoprotein/ABCB1 and down-regulates the expression and function. Cancer Lett 2014; 345: 124–131.
Yang M, Yuan ZM . A novel role of PRR14 in the regulation of skeletal myogenesis. Cell Death Dis 2015; 6: e1734.
Francis JC, Melchor L, Campbell J, Kendrick H, Wei W, Armisen-Garrido J et al. Whole-exome DNA sequence analysis of Brca2- and Trp53-deficient mouse mammary gland tumours. J Pathol 2015; 236: 186–200.
Reschen M, Kini U, Hood RL, Boycott KM, Hurst J, O’Callaghan CA . Floating-Harbor syndrome and polycystic kidneys associated with SRCAP mutation. Am J Med Genet A 2012; 158a: 3196–3200.
Liang X, Shan S, Pan L, Zhao J, Ranjan A, Wang F et al. Structural basis of H2A.Z recognition by SRCAP chromatin-remodeling subunit YL1. Nat Struct Mol Biol 2016; 23: 317–323.
Acknowledgements
Funding was provided by the German Cancer Aid, the Harald Huppert Foundations, The German Federal Ministry of Education and Research (eMed, Cliommics 01ZX1309B), the Multiple Myeloma Research Foundation, the Heinz Nixdorf Foundation (Germany), the Ministerium für Innovation, Wissenschaft und Forschung des Landes Nordrhein-Westfalen and the Faculty of Medicine University Duisburg-Essen. In the UK Myeloma UK and Bloodwise provided principal funding. Additional funding was provided by Cancer Research UK (C1298/A8362 supported by the Bobby Moore Fund) and The Rosetrees Trust. This study made use of genotyping data on the 1958 Birth Cohort generated by the Wellcome Trust Sanger Institute (http://www.wtccc.org.uk).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Leukemia website
Supplementary information
Rights and permissions
About this article
Cite this article
da Silva Filho, M., Försti, A., Weinhold, N. et al. Genome-wide association study of immunoglobulin light chain amyloidosis in three patient cohorts: comparison with myeloma. Leukemia 31, 1735–1742 (2017). https://doi.org/10.1038/leu.2016.387
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2016.387
This article is cited by
-
Genome-wide meta-analysis of monoclonal gammopathy of undetermined significance (MGUS) identifies risk loci impacting IRF-6
Blood Cancer Journal (2022)
-
Genetic pathogenesis of immunoglobulin light chain amyloidosis: basic characteristics and clinical applications
Experimental Hematology & Oncology (2021)
-
Systemic amyloidosis: moving into the spotlight
Leukemia (2020)
-
Genetic predisposition for multiple myeloma
Leukemia (2020)
-
Eight novel loci implicate shared genetic etiology in multiple myeloma, AL amyloidosis, and monoclonal gammopathy of unknown significance
Leukemia (2020)