Abstract
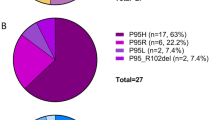
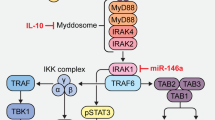
Myeloproliferative neoplasms (MPNs) feature a malignant clone containing the JAK2 V617F mutation, or another mutation causing dysregulated JAK2 kinase activity. The multiple disease phenotypes of MPNs, and their tendency to transform phenotypically, suggest pathophysiologic heterogeneities beyond a common phenomenon of JAK2 hyperactivation. JAK2 has the potential to activate multiple other signaling molecules, either directly through downstream effectors, or indirectly through induction of target gene expression. We have interrogated myeloproliferative signaling in myelofibrosis (MF) and secondary acute myeloid leukemia (sAML) patient samples using mass cytometry, which allows the quantitative measurement of multiple signaling molecules simultaneously at the single-cell level, in cell populations representing a nearly complete spectrum of hematopoiesis. MF and sAML malignant cells demonstrated a high prevalence of hyperactivation of the JAK-STAT, MAP kinase, PI3 kinase and NFκB signaling pathways. Constitutive NFκB signaling was evident across MF and sAML patients. A supporting gene set enrichment analysis (GSEA) of MF showed many NFκB target genes to be expressed above normal levels in MF patient CD34+ cells. NFκB inhibition suppressed colony formation from MF CD34+ cells. This study indicates that NFκB signaling contributes to human myeloproliferative disease and is abnormally activated in MF and sAML.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Hobbs GS, Rampal RK . Clinical and molecular genetic characterization of myelofibrosis. Curr Opin Hematol 2015; 22: 177–183.
Viny AD, Levine RL . Genetics of myeloproliferative neoplasms. Cancer J 2014; 20: 61–65.
Chen E, Beer PA, Godfrey AL, Ortmann CA, Li J, Costa-Pereira AP et al. Distinct clinical phenotypes associated with JAK2V617F reflect differential STAT1 signaling. Cancer Cell 2010; 18: 524–535.
Bendall SC, Simonds EF, Qiu P, Amir el AD, Krutzik PO, Finck R et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science 2011; 332: 687–696.
Bodenmiller B, Zunder ER, Finck R, Chen TJ, Savig ES, Bruggner RV et al. Multiplexed mass cytometry profiling of cellular states perturbed by small-molecule regulators. Nat Biotechnol 2012; 30: 858–867.
Amir, el AD, Davis KL, Tadmor MD, Simonds EF, Levine JH et al. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat Biotechnol 2013; 31: 545–552.
Behbehani GK, Samusik N, Bjornson ZB, Fantl WJ, Medeiros BC, Nolan GP . Mass cytometric functional profiling of acute myeloid leukemia defines cell-cycle and immunophenotypic properties that correlate with known responses to therapy. Cancer Discov 2015; 5: 988–1003.
Levine JH, Simonds EF, Bendall SC, Davis KL, Amir el AD, Tadmor MD et al. Data-driven phenotypic dissection of AML reveals progenitor-like cells that correlate with prognosis. Cell 2015; 162: 184–197.
Qiu P, Simonds EF, Bendall SC, Gibbs Jr KD, Bruggner RV, Linderman MD et al. Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nat Biotechnol 2011; 29: 886–891.
Irish JM, Hovland R, Krutzik PO, Perez OD, Bruserud O, Gjertsen BT et al. Single cell profiling of potentiated phospho-protein networks in cancer cells. Cell 2004; 118: 217–228.
Kotecha N, Flores NJ, Irish JM, Simonds EF, Sakai DS, Archambeault S et al. Single-cell profiling identifies aberrant STAT5 activation in myeloid malignancies with specific clinical and biologic correlates. Cancer Cell 2008; 14: 335–343.
Guzman ML, Neering SJ, Upchurch D, Grimes B, Howard DS, Rizzieri DA et al. Nuclear factor-kappaB is constitutively activated in primitive human acute myelogenous leukemia cells. Blood 2001; 98: 2301–2307.
Grosjean-Raillard J, Ades L, Boehrer S, Tailler M, Fabre C, Braun T et al. Flt3 receptor inhibition reduces constitutive NFkappaB activation in high-risk myelodysplastic syndrome and acute myeloid leukemia. Apoptosis 2008; 13: 1148–1161.
Grosjean-Raillard J, Tailler M, Ades L, Perfettini JL, Fabre C, Braun T et al. ATM mediates constitutive NF-kappaB activation in high-risk myelodysplastic syndrome and acute myeloid leukemia. Oncogene 2009; 28: 1099–1109.
Oh ST, Simonds EF, Jones C, Hale MB, Goltsev Y, Gibbs KD Jr et al. Novel mutations in the inhibitory adaptor protein LNK drive JAK-STAT signaling in patients with myeloproliferative neoplasms. Blood 2010; 116: 988–992.
Baxter EJ, Scott LM, Campbell PJ, East C, Fourouclas N, Swanton S et al. Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet 2005; 365: 1054–1061.
James C, Ugo V, Le Couedic JP, Staerk J, Delhommeau F, Lacout C et al. A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature 2005; 434: 1144–1148.
Kralovics R, Passamonti F, Buser AS, Teo SS, Tiedt R, Passweg JR et al. A gain-of-function mutation of JAK2 in myeloproliferative disorders. N Engl J Med 2005; 352: 1779–1790.
Levine RL, Wadleigh M, Cools J, Ebert BL, Wernig G, Huntly BJ et al. Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell 2005; 7: 387–397.
Schwemmers S, Will B, Waller CF, Abdulkarim K, Johansson P, Andreasson B et al. JAK2V617F-negative ET patients do not display constitutively active JAK/STAT signaling. Exp Hematol 2007; 35: 1695–1703.
Rampal R, Al-Shahrour F, Abdel-Wahab O, Patel JP, Brunel JP, Mermel CH et al. Integrated genomic analysis illustrates the central role of JAK-STAT pathway activation in myeloproliferative neoplasm pathogenesis. Blood 2014; 123: e123–e133.
Klampfl T, Gisslinger H, Harutyunyan AS, Nivarthi H, Rumi E, Milosevic JD et al. Somatic mutations of calreticulin in myeloproliferative neoplasms. N Engl J Med 2013; 369: 2379–2390.
Nangalia J, Massie CE, Baxter EJ, Nice FL, Gundem G, Wedge DC et al. Somatic CALR mutations in myeloproliferative neoplasms with nonmutated JAK2. N Engl J Med 2013; 369: 2391–2405.
Elf S, Abdelfattah NS, Chen E, Perales-Paton J, Rosen EA, Ko A et al. Mutant calreticulin requires both its mutant c-terminus and the thrombopoietin receptor for oncogenic transformation. Cancer Discov 2016; 6: 368–381.
Reuther GW . Recurring mutations in myeloproliferative neoplasms alter epigenetic regulation of gene expression. Am J Cancer Res 2011; 1: 752–762.
Abdel-Wahab O, Levine RL . Mutations in epigenetic modifiers in the pathogenesis and therapy of acute myeloid leukemia. Blood 2013; 121: 3563–3572.
Theocharides A, Boissinot M, Girodon F, Garand R, Teo SS, Lippert E et al. Leukemic blasts in transformed JAK2-V617F-positive myeloproliferative disorders are frequently negative for the JAK2-V617F mutation. Blood 2007; 110: 375–379.
Figueroa ME, Lugthart S, Li Y, Erpelinck-Verschueren C, Deng X, Christos PJ et al. DNA methylation signatures identify biologically distinct subtypes in acute myeloid leukemia. Cancer Cell 2010; 17: 13–27.
Kagoya Y, Yoshimi A, Kataoka K, Nakagawa M, Kumano K, Arai S et al. Positive feedback between NF-kappaB and TNF-alpha promotes leukemia-initiating cell capacity. J Clin Invest 2014; 124: 528–542.
Tefferi A, Vaidya R, Caramazza D, Finke C, Lasho T, Pardanani A . Circulating interleukin (IL)-8, IL-2 R, IL-12, and IL-15 levels are independently prognostic in primary myelofibrosis: a comprehensive cytokine profiling study. J Clin Oncol 2011; 29: 1356–1363.
Fleischman AG, Aichberger KJ, Luty SB, Bumm TG, Petersen CL, Doratotaj S et al. TNFalpha facilitates clonal expansion of JAK2V617F positive cells in myeloproliferative neoplasms. Blood 2011; 118: 6392–6398.
Levine RL, Belisle C, Wadleigh M, Zahrieh D, Lee S, Chagnon P et al. X-inactivation-based clonality analysis and quantitative JAK2V617F assessment reveal a strong association between clonality and JAK2V617F in PV but not ET/MMM, and identifies a subset of JAK2V617F-negative ET and MMM patients with clonal hematopoiesis. Blood 2006; 107: 4139–4141.
Engle EK, Fisher DA, Miller CA, McLellan MD, Fulton RS, Moore DM et al. Clonal evolution revealed by whole genome sequencing in a case of primary myelofibrosis transformed to secondary acute myeloid leukemia. Leukemia 2015; 29: 869–876.
Fienberg HG, Simonds EF, Fantl WJ, Nolan GP, Bodenmiller B . A platinum-based covalent viability reagent for single-cell mass cytometry. Cytometry A 2012; 81: 467–475.
Zunder ER, Finck R, Behbehani GK, Amir el AD, Krishnaswamy S, Gonzalez VD et al. Palladium-based mass tag cell barcoding with a doublet-filtering scheme and single-cell deconvolution algorithm. Nat Protoc 2015; 10: 316–333.
Behbehani GK, Thom C, Zunder ER, Finck R, Gaudilliere B, Fragiadakis GK et al. Transient partial permeabilization with saponin enables cellular barcoding prior to surface marker staining. Cytometry A 2014; 85: 1011–1019.
Majeti R, Park CY, Weissman IL . Identification of a hierarchy of multipotent hematopoietic progenitors in human cord blood. Cell Stem Cell 2007; 1: 635–645.
Krishnaswamy S, Spitzer MH, Mingueneau M, Bendall SC, Litvin O, Stone E et al. Systems biology. Conditional density-based analysis of T cell signaling in single-cell data. Science 2014; 346: 1250689.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005; 102: 15545–15550.
Norfo R, Zini R, Pennucci V, Bianchi E, Salati S, Guglielmelli P et al. miRNA-mRNA integrative analysis in primary myelofibrosis CD34+ cells: role of miR-155/JARID2 axis in abnormal megakaryopoiesis. Blood 2014; 124: e21–e32.
Kuo HP, Wang Z, Lee DF, Iwasaki M, Duque-Afonso J, Wong SH et al. Epigenetic roles of MLL oncoproteins are dependent on NF-kappaB. Cancer Cell 2013; 24: 423–437.
Gibbs Jr KD, Gilbert PM, Sachs K, Zhao F, Blau HM, Weissman IL et al. Single-cell phospho-specific flow cytometric analysis demonstrates biochemical and functional heterogeneity in human hematopoietic stem and progenitor compartments. Blood 2011; 117: 4226–4233.
Guglielmelli P, Zini R, Bogani C, Salati S, Pancrazzi A, Bianchi E et al. Molecular profiling of CD34+ cells in idiopathic myelofibrosis identifies a set of disease-associated genes and reveals the clinical significance of Wilms' tumor gene 1 (WT1). Stem Cells 2007; 25: 165–173.
Bosman MC, Schuringa JJ, Quax WJ, Vellenga E . Bortezomib sensitivity of acute myeloid leukemia CD34(+) cells can be enhanced by targeting the persisting activity of NF-kappaB and the accumulation of MCL-1. Exp Hematol 2013; 41: 530–538 e1.
Hariri F, Arguello M, Volpon L, Culjkovic-Kraljacic B, Nielsen TH, Hiscott J et al. The eukaryotic translation initiation factor eIF4E is a direct transcriptional target of NF-kappaB and is aberrantly regulated in acute myeloid leukemia. Leukemia 2013; 27: 2047–2055.
Wang X, Zhu K, Li S, Liao Y, Du R, Zhang X et al. MLL1, a H3K4 methyltransferase, regulates the TNFalpha-stimulated activation of genes downstream of NF-kappaB. J Cell Sci 2012; 125: 4058–4066.
Gleixner KV, Ferenc V, Peter B, Gruze A, Meyer RA, Hadzijusufovic E et al. Polo-like kinase 1 (Plk1) as a novel drug target in chronic myeloid leukemia: overriding imatinib resistance with the Plk1 inhibitor BI 2536. Cancer Res 2010; 70: 1513–1523.
Irish JM, Anensen N, Hovland R, Skavland J, Borresen-Dale AL, Bruserud O et al. Flt3 Y591 duplication and Bcl-2 overexpression are detected in acute myeloid leukemia cells with high levels of phosphorylated wild-type p53. Blood 2007; 109: 2589–2596.
Irish JM, Myklebust JH, Alizadeh AA, Houot R, Sharman JP, Czerwinski DK et al. B-cell signaling networks reveal a negative prognostic human lymphoma cell subset that emerges during tumor progression. Proc Natl Acad Sci USA 2010; 107: 12747–12754.
Neumann M, Naumann M . Beyond IkappaBs: alternative regulation of NF-kappaB activity. FASEB J 2007; 21: 2642–2654.
Nihira K, Ando Y, Yamaguchi T, Kagami Y, Miki Y, Yoshida K . Pim-1 controls NF-kappaB signalling by stabilizing RelA/p65. Cell Death Differ 2010; 17: 689–698.
Volk A, Li J, Xin J, You D, Zhang J, Liu X et al. Co-inhibition of NF-kappaB and JNK is synergistic in TNF-expressing human AML. J Exp Med 2014; 211: 1093–1108.
Rossi D, Ciardullo C, Gaidano G . Genetic aberrations of signaling pathways in lymphomagenesis: revelations from next generation sequencing studies. Semin Cancer Biol 2013; 23: 422–430.
Welch JS, Ley TJ, Link DC, Miller CA, Larson DE, Koboldt DC et al. The origin and evolution of mutations in acute myeloid leukemia. Cell 2012; 150: 264–278.
Delhommeau F, Dupont S, Tonetti C, Masse A, Godin I, Le Couedic JP et al. Evidence that the JAK2 G1849T (V617F) mutation occurs in a lymphomyeloid progenitor in polycythemia vera and idiopathic myelofibrosis. Blood 2007; 109: 71–77.
Acknowledgements
This work was supported by NIH grants K08HL106576 (Oh), K12HL087107 (Oh), and T32HL007088 (Engle, Fisher). This research was also supported by an American Cancer Society Postdoctoral Fellowship (Fisher). This work was also supported by a Doris Duke-Damon Runyon Clinical Investigator Award (Oh), BRIGHT Institute Pilot Research Project Award (Oh), Burroughs Wellcome Fund Collaborative Research Travel Grant (Oh), Sidney Kimmel Scholar Award (Oh), Leukemia Research Foundation New Investigator Award (Oh), and American Cancer Society Institutional Research Grant (Oh). Additional support was provided by the Washington University Institute of Clinical and Translational Sciences grant UL1TR000448 from the National Center for Advancing Translational Sciences of NIH. Support for patient sample collection and processing was provided by NIH grant P01CA101937. These studies were supported in part by funding provided by Incyte Corporation. Technical support was provided by the Alvin J Siteman Cancer Center Tissue Procurement Core Facility, Flow Cytometry Core, and Immunomonitoring Laboratory, which are supported by NCI Cancer Center Support Grant P30CA91842. The Immunomonitoring Laboratory is also supported by the Andrew M and Jane M Bursky Center for Human Immunology and Immunotherapy Programs. We thank C Miner for assistance with mass cytometry experiments, D Moore and K Luber for assistance with clinical samples and data, and D Bender and C Wilson for assistance with cytokine assays.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Leukemia website
Rights and permissions
About this article
Cite this article
Fisher, D., Malkova, O., Engle, E. et al. Mass cytometry analysis reveals hyperactive NF Kappa B signaling in myelofibrosis and secondary acute myeloid leukemia. Leukemia 31, 1962–1974 (2017). https://doi.org/10.1038/leu.2016.377
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2016.377
This article is cited by
-
miR-29b-3p suppresses the malignant biological behaviors of AML cells via inhibiting NF-κB and JAK/STAT signaling pathways by targeting HuR
BMC Cancer (2022)
-
Identification of potential pathways and microRNA-mRNA networks associated with benzene metabolite hydroquinone-induced hematotoxicity in human leukemia K562 cells
BMC Pharmacology and Toxicology (2022)
-
DUSP6 mediates resistance to JAK2 inhibition and drives leukemic progression
Nature Cancer (2022)
-
Paradigm shift: combination BET and JAK inhibition in myelofibrosis
Leukemia (2021)
-
Early and late stage MPN patients show distinct gene expression profiles in CD34+ cells
Annals of Hematology (2021)