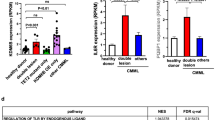
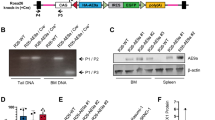
Abstract
TEN-ELEVEN-TRANSLOCATION-2 (TET2) and DNA-METHYLTRANSFERASE-3A (DNMT3A), both encoding proteins involved in regulating DNA methylation, are mutated in hematological malignancies affecting both myeloid and lymphoid lineages. We previously reported an association of TET2 and DNMT3A mutations in progenitors of patients with angioimmunoblastic T-cell lymphomas (AITL). Here, we report on the cooperative effect of Tet2 inactivation and DNMT3A mutation affecting arginine 882 (DNMT3AR882H) using a murine bone marrow transplantation assay. Five out of eighteen primary recipients developed hematological malignancies with one mouse developing an AITL-like disease, two mice presenting acute myeloid leukemia (AML)-like and two others T-cell acute lymphoblastic leukemia (T-ALL)-like diseases within 6 months following transplantation. Serial transplantations of DNMT3AR882H Tet2−/− progenitors led to a differentiation bias toward the T-cell compartment, eventually leading to AITL-like disease in 9/12 serially transplanted recipients. Expression profiling suggested that DNMT3AR882H Tet2−/− T-ALLs resemble those of NOTCH1 mutant. Methylation analysis of DNMT3AR882H Tet2−/− T-ALLs showed a global increase in DNA methylation affecting tumor suppressor genes and local hypomethylation affecting genes involved in the Notch pathway. Our data confirm the transformation potential of DNMT3AR882H Tet2−/− progenitors and represent the first cooperative model in mice involving Tet2 inactivation driving lymphoid malignancies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
References
Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med 2010; 363: 2424–2433.
Yan XJ, Xu J, Gu ZH, Pan CM, Lu G, Shen Y et al. Exome sequencing identifies somatic mutations of DNA methyltransferase gene DNMT3A in acute monocytic leukemia. Nat Genet 2011; 43: 309–315.
Couronne L, Bastard C, Bernard OA . TET2 and DNMT3A mutations in human T-cell lymphoma. N Engl J Med 2012; 366: 95–96.
Kim SJ, Zhao H, Hardikar S, Singh AK, Goodell MA, Chen T . A DNMT3A mutation common in AML exhibits dominant-negative effects in murine ES cells. Blood 2013; 122: 4086–4089.
Russler-Germain DA, Spencer DH, Young MA, Lamprecht TL, Miller CA, Fulton R et al. The R882H DNMT3A mutation associated with AML dominantly inhibits wild-type DNMT3A by blocking its ability to form active tetramers. Cancer Cell 2014; 25: 442–454.
Challen GA, Sun D, Jeong M, Luo M, Jelinek J, Berg JS et al. Dnmt3a is essential for hematopoietic stem cell differentiation. Nat Genet 2012; 44: 23–31.
Delhommeau F, Dupont S, Della Valle V, James C, Trannoy S, Masse et al. Mutation in TET2 in myeloid cancers. N Engl J Med 2009; 360: 2289–2301.
Quivoron C, Couronne L, Della Valle V, Lopez CK, Plo I, Wagner-Ballon O et al. TET2 inactivation results in pleiotropic hematopoietic abnormalities in mouse and is a recurrent event during human lymphomagenesis. Cancer Cell 2011; 20: 25–38.
Langemeijer SM, Kuiper RP, Berends M, Knops R, Aslanyan MG, Massop M et al. Acquired mutations in TET2 are common in myelodysplastic syndromes. Nat Genet 2009; 41: 838–842.
Scourzic L, Mouly E, Bernard OA . TET proteins and the control of cytosine demethylation in cancer. Genome Med 2015; 7: 9.
Chen E, Schneider RK, Breyfogle LJ, Rosen EA, Poveromo L, Elf S et al. Distinct effects of concomitant Jak2V617F expression and Tet2 loss in mice promote disease progression in myeloproliferative neoplasms. Blood 2015; 125: 327–335.
Kameda T, Shide K, Yamaji T, Kamiunten A, Sekine M, Taniguchi Y et al. Loss of TET2 has dual roles in murine myeloproliferative neoplasms: disease sustainer and disease accelerator. Blood 2015; 125: 304–315.
Lobry C, Ntziachristos P, Ndiaye-Lobry D, Oh P, Cimmino L, Zhu N et al. Notch pathway activation targets AML-initiating cell homeostasis and differentiation. J Exp Med 2013; 210: 301–319.
Muto T, Sashida G, Oshima M, Wendt GR, Mochizuki-Kashio M, Nagata Y et al. Concurrent loss of Ezh2 and Tet2 cooperates in the pathogenesis of myelodysplastic disorders. J Exp Med 2013; 210: 2627–2639.
Rasmussen KD, Jia G, Johansen JV, Pedersen MT, Rapin N, Bagger FO et al. Loss of TET2 in hematopoietic cells leads to DNA hypermethylation of active enhancers and induction of leukemogenesis. Genes Dev 2015; 29: 910–922.
Shih AH, Jiang Y, Meydan C, Shank K, Pandey S, Barreyro L et al. Mutational cooperativity linked to combinatorial epigenetic gain of function in acute myeloid leukemia. Cancer Cell 2015; 27: 502–515.
Soucie E, Hanssens K, Mercher T, Georgin-Lavialle S, Damaj G, Livideanu C et al. In aggressive forms of mastocytosis, TET2 loss cooperates with c-KITD816V to transform mast cells. Blood 2012; 120: 4846–4849.
Palomero T, Couronne L, Khiabanian H, Kim MY, Ambesi-Impiombato A, Perez-Garcia et al. Recurrent mutations in epigenetic regulators, RHOA and FYN kinase in peripheral T cell lymphomas. Nat Genet 2014; 46: 166–170.
Sakata-Yanagimoto M, Enami T, Yoshida K, Shiraishi Y, Ishii R, Miyake Y et al. Somatic RHOA mutation in angioimmunoblastic T cell lymphoma. Nat Genet 2014; 46: 171–175.
Lemonnier F, Couronne L, Parrens M, Jais JP, Travert M, Lamant L et al. Recurrent TET2 mutations in peripheral T-cell lymphomas correlate with TFH-like features and adverse clinical parameters. Blood 2012; 120: 1466–1469.
Malinge S, Ragu C, Della-Valle V, Pisani D, Constantinescu SN, Perez C et al. Activating mutations in human acute megakaryoblastic leukemia. Blood 2008; 112: 4220–4226.
Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PA, Rappsilber J et al. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature 2011; 473: 343–348.
Gu H, Smith ZD, Bock C, Boyle P, Gnirke A, Meissner A . Preparation of reduced representation bisulfite sequencing libraries for genome-scale DNA methylation profiling. Nat Protoc 2011; 6: 468–481.
Thiollier C, Lopez CK, Gerby B, Ignacimouttou C, Poglio S, Duffourd Y et al. Characterization of novel genomic alterations and therapeutic approaches using acute megakaryoblastic leukemia xenograft models. J Exp Med 2012; 209: 2017–2031.
Crotty S . Follicular helper CD4 T cells (TFH). Annu Rev Immunol 2011; 29: 621–663.
de Leval L, Rickman DS, Thielen C, Reynies A, Huang YL, Delsol G et al. The gene expression profile of nodal peripheral T-cell lymphoma demonstrates a molecular link between angioimmunoblastic T-cell lymphoma (AITL) and follicular helper T (TFH) cells. Blood 2007; 109: 4952–4963.
Virgilio L, Lazzeri C, Bichi R, Nibu K, Narducci MG, Russo G et al. Deregulated expression of TCL1 causes T cell leukemia in mice. Proc Natl Acad Sci USA 1998; 95: 3885–3889.
Chiang MY, Xu L, Shestova O, Histen G, L'Heureux S, Romany C et al. Leukemia-associated NOTCH1 alleles are weak tumor initiators but accelerate K-ras-initiated leukemia. J Clin Invest 2008; 118: 3181–3194.
Tsagaratou A, Aijo T, Lio CW, Yue X, Huang Y, Jacobsen SE et al. Dissecting the dynamic changes of 5-hydroxymethylcytosine in T-cell development and differentiation. Proc Natl Acad Sci USA 2014; 111: E3306–E3315.
Steinke FC, Yu S, Zhou X, He B, Yang W, Zhou B et al. TCF-1 and LEF-1 act upstream of Th-POK to promote the CD4(+) T cell fate and interact with Runx3 to silence Cd4 in CD8(+) T cells. Nat Immunol 2014 Jul; 15: 646–656.
Yu S, Xue HH . TCF-1 mediates repression of Notch pathway in T lineage-committed early thymocytes. Blood 2013; 121: 4008–4009.
Staal FJ, Clevers H . Tales of the unexpected: Tcf1 functions as a tumor suppressor for leukemias. Immunity 2012; 37: 761–763.
Li Chew C, Lunardi A, Gulluni F, Ruan DT, Chen M, Salmena L et al. In vivo role of INPP4B in tumor and metastasis suppression through regulation of PI3K-AKT signaling at endosomes. Cancer Discov 2015; 5: 740–751.
Kofuji S, Kimura H, Nakanishi H, Nanjo H, Takasuga S, Liu H et al. INPP4B Is a PtdIns(3,4,5)P3 phosphatase that can act as a tumor suppressor. Cancer Discov 2015; 5: 730–739.
Greif PA, Eck SH, Konstandin NP, Benet-Pages A, Ksienzyk B, Dufour et al. Identification of recurring tumor-specific somatic mutations in acute myeloid leukemia by transcriptome sequencing. Leukemia 2011; 25: 821–827.
Wei J, Wunderlich M, Fox C, Alvarez S, Cigudosa JC, Wilhelm JS et al. Microenvironment determines lineage fate in a human model of MLL-AF9 leukemia. Cancer Cell 2008; 13: 483–495.
Mayle A, Yang L, Rodriguez B, Zhou T, Chang E, Curry CV et al. Dnmt3a loss predisposes murine hematopoietic stem cells to malignant transformation. Blood 2015; 125: 629–638.
Peters SL, Hlady RA, Opavska J, Klinkebiel D, Pirruccello SJ, Talmon GA et al. Tumor suppressor functions of Dnmt3a and Dnmt3b in the prevention of malignant mouse lymphopoiesis. Leukemia 2014; 28: 1138–1142.
Dorfman DM, Greisman HA, Shahsafaei A . Loss of expression of the WNT/beta-catenin-signaling pathway transcription factors lymphoid enhancer factor-1 (LEF-1) and T cell factor-1 (TCF-1) in a subset of peripheral T cell lymphomas. Am J Pathol 2003; 162: 1539–1544.
Lee HO, He X, Mookerjee-Basu J, Zhongping D, Hua X, Nicolas E et al. Disregulated expression of the transcription factor ThPOK during T-cell development leads to high incidence of T-cell lymphomas. Proc Natl Acad Sci USA 2015; 112: 7773–7778.
Grossmann V, Haferlach C, Weissmann S, Roller A, Schindela S, Poetzinger F et al. The molecular profile of adult T-cell acute lymphoblastic leukemia: mutations in RUNX1 and DNMT3A are associated with poor prognosis in T-ALL. Genes Chromosomes Cancer 2013; 52: 410–422.
Neumann M, Heesch S, Schlee C, Schwartz S, Gokbuget N, Hoelzer D et al. Whole-exome sequencing in adult ETP-ALL reveals a high rate of DNMT3A mutations. Blood 2013; 121: 4749–4752.
Ellyard JI, Chia T, Rodriguez-Pinilla SM, Martin JL, Hu X, Navarro-Gonzalez M et al. Heterozygosity for Roquinsan leads to angioimmunoblastic T-cell lymphoma-like tumors in mice. Blood 2012; 120: 812–821.
Muto H, Sakata-Yanagimoto M, Nagae G, Shiozawa Y, Miyake Y, Yoshida K et al. Reduced TET2 function leads to T-cell lymphoma with follicular helper T-cell-like features in mice. Blood Cancer J 2014; 4: e264.
Kluk MJ, Ashworth T, Wang H, Knoechel B, Mason EF, Morgan EA et al. Gauging NOTCH1 activation in cancer using immunohistochemistry. PLoS One 2013; 8: e67306.
Broske AM, Vockentanz L, Kharazi S, Huska MR, Mancini E, Scheller M et al. DNA methylation protects hematopoietic stem cell multipotency from myeloerythroid restriction. Nat Genet 2009; 41: 1207–1215.
Auderset F, Schuster S, Fasnacht N, Coutaz M, Charmoy M, Koch U et al. Notch signaling regulates follicular helper T cell differentiation. J Immunol 2013; 191: 2344–2350.
Fasnacht N, Huang HY, Koch U, Favre S, Auderset F, Chai Q et al. Specific fibroblastic niches in secondary lymphoid organs orchestrate distinct Notch-regulated immune responses. J Exp Med 2014; 211: 2265–2279.
Acknowledgements
We thank members of the Bernard laboratory for helpful discussions, Julie Mouillaux for her involvement in this project, Patrick Gonin from the Gustave Roussy animal facility for excellent mouse care as well as Yann Lecluse and Philippe Rameau from the Gustave Roussy Flow Cytometry Core Facility. We also thank the Gustave Roussy Genomic Platform for high-throughput sequencing. Work in the laboratory was supported by grants from INSERM, Institut National du Cancer (INCA), 2013-1-PL BIO-09, INCa-DGOS-INSERM 6043, Ligue Nationale Contre le Cancer (LNCC), Fondation pour la Recherche Médicale (FRM) and Association Laurette Fugain. The work in the laboratory of KH was supported by the Danish Cancer Society, the European Research Council (294666_DNAMET), the Danish National Research Foundation (DNRF82) and the Novo Nordisk Foundation. LS is supported by fellowships from Cancéropôle Ile de France and Fondation Association pour la recherche sur le Cancer (ARC).
Author contributions
LS drafted the manuscript. LS and LC designed, performed and analyzed results. VDV, ElM, JC, EnM, CkL NM, ND, PG, MW, FP, MP, AB and KH performed experiments and analyzed data. MBD, PhD, MF, SG and PaD analyzed results. TM designed experiments, analyzed results and drafted the manuscript. OAB designed the project, experiments and wrote the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Leukemia website
Rights and permissions
About this article
Cite this article
Scourzic, L., Couronné, L., Pedersen, M. et al. DNMT3AR882H mutant and Tet2 inactivation cooperate in the deregulation of DNA methylation control to induce lymphoid malignancies in mice. Leukemia 30, 1388–1398 (2016). https://doi.org/10.1038/leu.2016.29
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2016.29
This article is cited by
-
DNMT3AR882H accelerates angioimmunoblastic T-cell lymphoma in mice
Oncogene (2023)
-
Advances in the pathogenesis and therapeutic strategies of angioimmunoblastic T-cell lymphoma
Clinical and Experimental Medicine (2023)
-
Mature T-cell and NK-cell lymphomas: updates on molecular genetic features
International Journal of Hematology (2023)
-
Peripheral T cell lymphomas: from the bench to the clinic
Nature Reviews Cancer (2020)
-
New preclinical models for angioimmunoblastic T-cell lymphoma: filling the GAP
Oncogenesis (2020)