Abstract
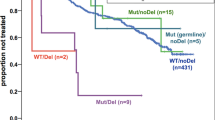
Alterations in TP53 have been described in many cancer types including hematological neoplasms. We aimed at comparing TP53 mutations (mut) and deletions (del) in a large cohort of patients with hematological malignancies (n=3307), including AML (n=858), MDS (n=943), ALL (n=358), CLL (n=1148). Overall, alterations in TP53 were detected in 332/3307 cases (10%). The highest frequency was observed in ALL (total: 19%; mut+del: 6%; mut only: 8%; del only: 5%) and AML (total: 13%; mut+del: 5%; mut only: 7%; del only: 1%), whereas TP53 alterations occurred less frequently in CLL (total: 8%) and MDS (total: 7%). TP53 mutations were significantly more frequent in patients ⩾60 vs <60 years in AML (9% vs 2%, P<0.001) and ALL (12% vs 6%, P<0.001). TP53mut+del had a significant negative impact on overall survival in all entities, whereas differences were observed regarding TP53mut only or TP53del only: TP53mut only impacted survival in AML (36 vs 9 months, P<0.001) and MDS (65 vs 19 months, P<0.001), TP53del only in CLL (not reached vs 64 months, P=0.008) and MDS (65 vs 24 months, P=0.011). As substantial differences between the entities are observed regarding correlation to age and survival, we suggest evaluation of both TP53 deletion and mutation status.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Rivlin N, Brosh R, Oren M, Rotter V . Mutations in the p53 Tumor Suppressor Gene: Important Milestones at the Various Steps of Tumorigenesis. Genes Cancer 2011; 2: 466–474.
Preudhomme C, Fenaux P . The clinical significance of mutations of the P53 tumour suppressor gene in haematological malignancies. Br J Haematol 1997; 98: 502–511.
Soussi T, Legros Y, Lubin R, Ory K, Schlichtholz B . Multifactorial analysis of p53 alteration in human cancer: A review. Int J Cancer 1994; 57: 1–9.
Wickremasinghe RG, Prentice AG, Steele AJ . p53 and Notch signaling in chronic lymphocytic leukemia: clues to identifying novel therapeutic strategies. Leukemia 2011; 25: 1400–1407.
Lane DP . Cancer. p53, guardian of the genome. Nature 1992; 358: 15–16.
Fenaux P, Preudhomme C, Quiquandon I, Jonveaux P, Laï JL, Vanrumbeke M et al. Mutations of the P53 gene in acute myeloid leukaemia. Br J Haematol 1992; 80: 178–183.
Zenz T, Eichhorst B, Busch R, Denzel T, Häbe S, Winkler D et al. TP53 mutation and survival in chronic lymphocytic leukemia. J Clin Oncol 2010; 28: 4473–4479.
Rotter V, Aloni-Grinstein R, Schwartz D, Elkind NB, Simons A, Wolkowicz R et al. Does wild-type p53 play a role in normal cell differentiation? Semin Cancer Biol 1994; 5: 229–236.
Wynford-Thomas D . Cellular senescence and cancer. J Pathol 1999; 187: 100–111.
Agirre X, Novo FJ, Calasanz MJ, Larráyoz MJ, Lahortiga I, Valgañón M et al. TP53 is frequently altered by methylation, mutation, and/or deletion in acute lymphoblastic leukaemia. Mol Carcinog 2003; 38: 201–208.
Knudson AG . Mutation and Cancer: Statistical Study of Retinoblastoma. Proc Natl Acad of Sci USA 1971; 68: 820–823.
Venot C, Maratrat M, Dureuil C, Conseiller E, Bracco L, Debussche L . The requirement for the p53 proline-rich functional domain for mediation of apoptosis is correlated with specific PIG3 gene transactivation and with transcriptional repression. EMBO J 1998; 17: 4668–4679.
Harms KL, Chen X . The C terminus of p53 Family Proteins Is a Cell Fate Determinant. Mol Cell Biol 2005; 25: 2014–2030.
Shaw P, Freeman J, Bovey R, Iggo R . Regulation of specific DNA binding by p53: Evidence for a role for O-glycosylation and charged residues at the carboxy-terminus. Oncogene 1996; 12: 921–930.
Sakaguchi K, Herrera JE, Saito S, Miki T, Bustin M, Vassilev A et al. DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev 1998; 12: 2831–2841.
Kruse JP, Gu W . Modes of p53 regulation. Cell 2009; 137: 609–622.
Rossi D, Gaidano G . Molecular Genetics of High-risk Chronic Lymphocytic Leukemia. Expert Rev Hematol 2012; 5: 593–602.
Hollstein M, Rice K, Greenblatt MS, Soussi T, Fuchs R, Sorlie T et al. Database of p53 gene somatic mutations in human tumors and cell lines. Nucleic Acids Res 1994; 22: 3551–3555.
Bullock AN, Fersht AR . Rescuing the function of mutant p53. Nat Rev Cancer 2001; 1: 68–76.
Brosh R, Rotter V . When mutants gain new powers: news from the mutant p53 field. Nat Rev Cancer 2009; 9: 701–713.
Li J, Yang L, Gaur S, Zhang K, Wu X, Yuan YC et al. Mutants TP53 p.R273H and p.R273C but not p.R273G enhance cancer cell malignancy. Hum Mutat 2014; 35: 575–584.
Stengel A, Schnittger S, Weissmann S, Kuznia S, Kern W, Kohlmann A et al. TP53 mutations occur in 15.7% of ALL and are associated with MYC-rearrangement, low hypodiploidy, and a poor prognosis. Blood 2014; 124: 251–258.
Grossmann V, Schnittger S, Kohlmann A, Eder C, Roller A, Dicker F et al. A novel hierarchical prognostic model of AML solely based on molecular mutations. Blood 2012; 120: 2963–2972.
Rücker FG, Schlenk RF, Bullinger L, Kayser S, Teleanu V, Kett H et al. TP53 alterations in acute myeloid leukemia with complex karyotype correlate with specific copy number alterations, monosomal karyotype, and dismal outcome. Blood 2012; 119: 2114–2121.
Döhner H, Stilgenbauer S, Benner A, Leupolt E, Kröber A, Bullinger L et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med 2000; 343: 1910–1916.
Rossi D, Cerri M, Deambrogi C, Sozzi E, Cresta S, Rasi S et al. The prognostic value of TP53 mutations in chronic lymphocytic leukemia is independent of Del17p13: implications for overall survival and chemorefractoriness. Clin Cancer Res 2009; 15: 995–1004.
Jeromin S, Weissmann S, Haferlach C, Dicker F, Bayer K, Grossmann V et al. SF3B1 mutations correlated to cytogenetics and mutations in NOTCH1, FBXW7, MYD88, XPO1 and TP53 in 1160 untreated CLL patients. Leukemia 2014; 28: 108–117.
Cazzola M, Della Porta MG, Malcovati L . The genetic basis of myelodysplasia and its clinical relevance. Blood 2013; 122: 4021–4034.
Papaemmanuil E, Gerstung M, Malcovati L, Tauro S, Gundem G, Van Loo P et al. Chronic Myeloid Disorders Working Group of the International Cancer Genome Consortium. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood 2013; 122: 3616–3627.
Haferlach T, Nagata Y, Grossmann V, Okuno Y, Bacher U, Nagae G et al. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia 2014; 28: 241–247.
Hof J, Krentz S, van Schewick C, Körner G, Shalapour S, Rhein P et al. Mutations and deletions of the TP53 gene predict nonresponse to treatment and poor outcome in first relapse of childhood acute lymphoblastic leukemia. J Clin Oncol 2011; 29: 3185–3193.
Wada M, Bartram CR, Nakamura H, Hachiya M, Chen DL, Borenstein J et al. Analysis of p53 mutations in a large series of lymphoid hematologic malignancies of childhood. Blood 1993; 82: 3163–3169.
Wong TN, Ramsingh G, Young AL, Miller CA, Touma W, Welch JS et al. Role of TP53 mutations in the origin and evolution of therapy-related acute myeloid leukaemia. Nature 2015; 518: 552–555.
Haferlach T, Kern W, Schoch C, Hiddemann W, Sauerland MC . Morphologic dysplasia in acute myeloid leukemia: importance of granulocytic dysplasia. J Clin Oncol 2003; 21: 3004–3005.
Kern W, Voskova D, Schoch C, Hiddemann W, Schnittger S, Haferlach T . Determination of relapse risk based on assessment of minimal residual disease during complete remission by multiparameter flow cytometry in unselected patients with acute myeloid leukemia. Blood 2004; 104: 3078–3085.
Bene MC, Castoldi G, Knapp W, Ludwig WD, Matutes E, Orfao A et al. Proposals for the immunological classification of acute leukemias. European Group for the Immunological Characterization of Leukemias (EGIL). Leukemia 1995; 9: 1783–1786.
Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H et al WHO Classification of Tumours of Haematopoietic and Lympoid Tissues. IARC: Lyon, France, 2008.
Schoch C, Schnittger S, Bursch S, Gerstner D, Hochhaus A, Berger U et al. Comparison of chromosome banding analysis, interphase- and hypermetaphase-FISH, qualitative and quantitative PCR for diagnosis and for follow-up in chronic myeloid leukemia: a study on 350 cases. Leukemia 2002; 16: 53–59.
Dicker F, Schnittger S, Haferlach T, Kern W, Schoch C . Immunostimulatory oligonucleotide-induced metaphase cytogenetics detect chromosomal aberrations in 80% of CLL patients: A study of 132 CLL cases with correlation to FISH, IgVH status, and CD38 expression. Blood 2006; 108: 3152–3160.
Haferlach C, Bacher U . Cytogenetic methods in chronic lymphocytic leukemia. Methods Mol Biol 2011; 730: 119–130.
Shaffer LG, Tommerup N . ISCN 2013: an International System for Human Cytogenetic Nomenclature. Karger: Basel, New York, 2013.
Petitjean A, Mathe E, Kato S, Ishioka C, Tavtigian SV, Hainaut P et al. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum Mutat 2007; 28: 622–629.
Ashour Ahmed A, Etemadmoghadam D, Temple J, Lynch AG, Riad M, Sharma R et al. Brenton Driver mutations in TP53 are ubiquitous in high grade serous carcinoma of the ovary. J Pathol 2010; 221: 49–56.
O’Hara AJ, Bell DW . The genomics and genetics of endometrial cancer. Adv Genomics Genet 2012; 2012: 33–47.
Stilgenbauer S, Schnaiter A, Paschka P, Zenz T, Rossi M, Döhner K et al. Gene mutations and treatment outcome in chronic lymphocytic leukemia: results from the CLL8 trial. Blood 2014; 123: 3247–3254.
Tam CS, Stilgenbauer S . How best to manage patients with chronic lymphocytic leuekmia with 17p deletion and/or TP53 mutation? Leuk Lymphoma 2015; 56: 587–593.
Bejar R, Stevenson KE, Caughey B, Lindsley RC, Mar BG, Stojanov P et al. Somatic mutations predict poor outcome in patients with myelodysplastic syndrome after hematopoietic stem-cell transplantation. J Clin Oncol 2014; 32: 2691–2698.
Jadersten M, Saft L, Smith A, Kulasekararaj A, Pomplun S, Göhring G et al. TP53 mutations in low-risk myelodysplastic syndromes with del(5q) predict disease progression. J Clin Oncol 2011; 29: 1971–1979.
Xu J, Wang J, Hu Y, Qian J, Xu B, Chen H et al. Unequal prognostic potentials of p53 gain-of-function mutations in human cancers associate with drug-metabolizing activity. Cell Death Dis 2014; 5: e1108.
Joerger AC, Fersht AR . The tumor suppressor p53: from structures to drug discovery. Cold Spring Harb Perspect Biol 2010; 2: a000919.
Haferlach C, Dicker F, Herholz H, Schnittger S, Kern W, Haferlach T . Mutations of the TP53 gene in acute myeloid leukemia are strongly associated with a complex aberrant karyotype. Leukemia 2008; 22: 1539–1541.
Fernandez-Mercado M, Burns A, Pellagatti A, Giagounidis A, Germing U, Agirre X et al. Targeted re-sequencing analysis of 25 genes commonly mutated in myeloid disorders in del(5q) myelodysplastic syndromes. Haematologica 2013; 98: 1856–1864.
Rossi D, Khiabanian H, Spina V, Ciardullo C, Bruscaggin A, Famà R et al. Clinical impact of small TP53 mutated subclones in chronic lymphocytic leukemia. Blood 2014; 123: 2139–2147.
Acknowledgements
We thank all co-workers at the MLL Munich Leukemia Laboratory for their technical assistance. We thank all physicians for providing and caring for patients as well as collecting the data.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
CH, WK and TH declare part ownership of MLL Munich Leukemia Laboratory. AS, MM and AF are employed by MLL Munich Leukemia Laboratory.
Additional information
Supplementary Information accompanies this paper on the Leukemia website
Rights and permissions
About this article
Cite this article
Stengel, A., Kern, W., Haferlach, T. et al. The impact of TP53 mutations and TP53 deletions on survival varies between AML, ALL, MDS and CLL: an analysis of 3307 cases. Leukemia 31, 705–711 (2017). https://doi.org/10.1038/leu.2016.263
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2016.263
This article is cited by
-
Prevalence, causes and impact of TP53-loss phenocopying events in human tumors
BMC Biology (2023)
-
Accurate interpretation of p53 immunohistochemical patterns is a surrogate biomarker for TP53 alterations in large B-cell lymphoma
BMC Cancer (2023)
-
Combined inhibition of BCL-2 and MCL-1 overcomes BAX deficiency-mediated resistance of TP53-mutant acute myeloid leukemia to individual BH3 mimetics
Blood Cancer Journal (2023)
-
Somatic TP53 single nucleotide variants, indels and copy number alterations in chronic myelomonocytic leukemia (CMML)
Leukemia (2023)
-
Deep genomic characterization highlights complexities and prognostic markers of pediatric acute myeloid leukemia
Communications Biology (2023)