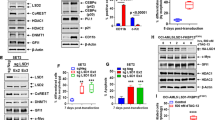
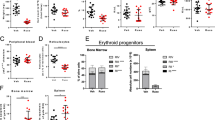
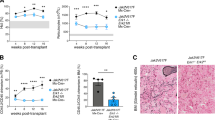
Abstract
Myeloproliferative neoplasms with myelofibrosis (MPN-MF) demonstrate constitutive activation of Janus kinase/signal transducer and activator of transcription (JAK/STAT) signaling that responds to treatment with the JAK1 and 2 kinase inhibitor (JAKi) ruxolitinib. However, MPN-MF often progresses (~20%) to secondary acute myeloid leukemia (sAML), where standard induction chemotherapy or ruxolitinib is relatively ineffective, necessitating the development of novel therapeutic approaches. In the present studies, we demonstrate that treatment with BET (bromodomain and extraterminal) protein inhibitor (BETi), for example, JQ1, inhibits growth and induces apoptosis of cultured and primary, patient-derived (PD), post-MPN sAML blast progenitor cells. Reverse-phase protein array, mass-cytometry and Western analyses revealed that BETi treatment attenuated the protein expressions of c-MYC, p-STAT5, Bcl-xL, CDK4/6, PIM1 and IL-7R, whereas it concomitantly induced the levels of HEXIM1, p21 and BIM in the sAML cells. Co-treatment with BETi and ruxolitinib synergistically induced apoptosis of cultured and PD sAML cells, as well as significantly improved survival of immune-depleted mice engrafted with human sAML cells. Although BETi or heat shock protein 90 inhibitor (HSP90i) alone exerted lethal activity, cotreatment with BETi and HSP90i was synergistically lethal against the ruxolitinib-persister or ruxolitinib-resistant sAML cells. Collectively, these findings further support in vivo testing of BETi-based combinations with JAKi and HSP90i against post-MPN sAML cells.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Vainchenker W, Delhommeau F, Constantinescu SN, Bernard OA . New mutations and pathogenesis of myeloproliferative neoplasms. Blood 2011; 118: 1723–1735.
Rampal R, Al-Shahrour F, Abdel-Wahab O, Patel JP, Brunel JP, Mermel CH et al. Integrated genomic analysis illustrates the central role of JAK-STAT pathway activation in myeloproliferative neoplasm pathogenesis. Blood 2014; 123: e123–e133.
Keohane C, Mesa R, Harrison C . The role of JAK1/2 inhibitors in the treatment of chronic myeloproliferative neoplasms. Am Soc Clin Oncol Educ Book 2013, 301–305.
Vannucchi AM, Kiladjian JJ, Griesshammer M, Masszi T, Durrant S, Passamonti F et al. Ruxolitinib versus standard therapy for the treatment of polycythemia vera. N Engl J Med 2015; 372: 426–435.
Vannucchi AM, Kantarjian HM, Kiladjian JJ, Gotlib J, Cervantes F, Mesa RA et al. A pooled analysis of overall survival in COMFORT-I and COMFORT-II, 2 randomized phase III trials of ruxolitinib for the treatment of myelofibrosis. Haematologica 2015; 100: 1139–1145.
Passamonti F, Maffioli M, Cervantes F, Vannucchi AM, Morra E, Barbui T et al. Impact of ruxolitinib on the natural history of primary myelofibrosis: a comparison of the DIPSS and the COMFORT-2 cohorts. Blood 2014; 123: 1833–1835.
Guglielmelli P, Biamonte F, Rotunno G, Artusi V, Artuso L, Bernardis I et al. Impact of mutational status on outcomes in myelofibrosis patients treated with ruxolitinib in the COMFORT-II study. Blood 2014; 123: 2157–2160.
Fiskus W, Verstovsek S, Manshouri T, Rao R, Balusu R, Venkannagari S et al. Heat shock protein 90 inhibitor is synergistic with JAK2 inhibitor and overcomes resistance to JAK2-TKI in human myeloproliferative neoplasm cells. Clin Cancer Res 2011; 17: 7347–7358.
Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell 2010; 141: 69–80.
Koppikar P, Bhagwat N, Kilpivaara O, Manshouri T, Adli M, Hricik T et al. Heterodimeric JAK-STAT activation as a mechanism of persistence to JAK2 inhibitor therapy. Nature 2012; 489: 155–159.
Meyer SC, Levine RL . Molecular pathways: molecular basis for sensitivity and resistance to JAK kinase inhibitors. Clin Cancer Res 2014; 20: 2051–2059.
Lundberg P, Karow A, Nienhold R, Looser R, Hao-Shen H, Nissen I et al. Clonal evolution and clinical correlates of somatic mutations in myeloproliferative neoplasms. Blood 2014; 123: 2220–2228.
Hobbs GS, Rampal RK . Clinical and molecular genetic characterization of myelofibrosis. Curr Opin Hematol 2015; 22: 177–183.
Lasho TL, Jimma T, Finke CM, Patnaik M, Hanson CA, Ketterling RP et al. SRSF2 mutations in primary myelofibrosis: significant clustering with IDH mutations and independent association with inferior overall and leukemia-free survival. Blood 2012; 120: 4168–4171.
Zhang SJ, Rampal R, Manshouri T, Patel J, Mensah N, Kayserian A et al. Genetic analysis of patients with leukemic transformation of myeloproliferative neoplasms shows recurrent SRSF2 mutations that are associated with adverse outcome. Blood 2012; 119: 4480–4485.
Rampal R, Mascarenhas J . Pathogenesis and management of acute myeloid leukemia that has evolved from a myeloproliferative neoplasm. Curr Opin Hematol 2014; 21: 65–71.
Eghtedar A, Verstovsek S, Estrov Z, Burger J, Cortes J, Bivins C et al. Phase 2 study of the JAK kinase inhibitor ruxolitinib in patients with refractory leukemias, including postmyeloproliferative neoplasm acute myeloid leukemia. Blood 2012; 119: 4614–4618.
Kundranda MN, Tibes R, Mesa RA . Transformation of a chronic myeloproliferative neoplasm to acute myelogenous leukemia: does anything work? Curr Hematol Malig Rep 2012; 7: 78–86.
Rampal R, Ahn J, Abdel-Wahab O, Nahas M, Wang K, Lipson D et al. Genomic and functional analysis of leukemic transformation of myeloproliferative neoplasms. Proc Natl Acad Sci USA 2014; 111: E5401–E5410.
Milosevic JD, Puda A, Malcovati L, Berg T, Hofbauer M, Stukalov A et al. Clinical significance of genetic aberrations in secondary acute myeloid leukemia. Am J Hematol 2012; 87: 1010–1016.
Shi J, Vakoc CR . The mechanism behind the therapeutic activity of BET bromodomain inhibition. Mol Cell 2014; 54: 728–736.
Roe JS, Mercan F, Rivera K, Pappin DJ, Vakoc CR . BET bromodomain inhibition suppresses the function of hematopoietic transcription factors in acute myeloid leukemia. Mol Cell 2015; 58: 1028–1039.
Itzen F, Greifenberg AK, Bosken CA, Geyer M . Brd4 activates P-TEFB for RNA polymerase II CTD phosphorylation. Nucleic Acids Res 2014; 42: 7577–7590.
Nechaev S, Adelman K . Pol II waiting in the starting gates: regulating the transition from transcription initiation into productive elongation. Biochim Biophys Acta 2011; 1809: 34–45.
Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR . Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 2013; 153: 320–334.
Bolden JE, Tasdemir N, Dow LE, van Es JH, Wilkinson JE, Zhao Z et al. Inducible in vivo silencing of Brd4 identifies potential toxicities of sustained BET protein inhibition. Cell Rep 2014; 8: 1919–1929.
Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature 2011; 478: 524–528.
Filippakapoulos P, Knapp S . Targeting bromodomains: epigenetic readers of lysine acetylation. Nat Rev Drug Discov 2014; 13: 337–356.
Dawson MA, Prinjha RK, Dittmann A, Giotopoulos G, Bantscheff M, Chan WI et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature 2011; 478: 529–533.
Fiskus W, Sharma S, Qi J, Valenta JA, Schaub LJ, Shah B et al. Highly active combination of BRD4 antagonist and histone deacetylase inhibitor against human acute myelogenous leukemia cells. Mol Cancer Ther 2014; 13: 1142–1154.
Dawson MA, Gudgin EJ, Horton SJ, Giotopoulos G, Meduri E, Robson S et al. Recurrent mutations, including NPM1c, activate a BRD4-dependent core transcriptional program in acute myeloid leukemia. Leukemia 2014; 28: 311–320.
Fiskus W, Sharma S, Qi J, Shah B, Devaraj SG, Leveque C et al. BET protein antagonist JQ1 is synergistically lethal with FLT3 tyrosine kinase inhibitor (TKI) and overcomes resistance to FLT3-TKI in AML cells expressing FLT-ITD. Mol Cancer Ther 2014; 13: 2315–2327.
Devaraj SG, Fiskus W, Shah B, Qi J, Sun B, Iyer SP et al. HEXIM1 induction is mechanistically involved in mediating anti-AML activity of BET protein bromodomain antagonist. Leukemia 2016; 30: 504–508.
Sonkin D, Palmer M, Rong X, Horrigan K, Regnier CH, Fanton C et al. The identification and characterization of a STAT5 gene signature in hematologic malignancies. Cancer Biomark 2015; 15: 79–87.
Ott CJ, Kopp N, Bird L, Paranal RM, Qi J, Bowman T et al. BET bromodomain inhibition targets both c-Myc and IL7R in high-risk acute lymphoblastic leukemia. Blood 2012; 120: 2843–2852.
Sun B, Shah B, Fiskus W, Qi J, Rajapakshe K, Coarfa C et al. Synergistic activity of BET protein bromodomain antagonist-based combinations against Mantle Cell Lymphoma (MCL) cells sensitive or resistant to ibrutinib. Blood 2015; 126: 1565–1574.
Hart S, Goh KC, Novotny-Diermayr V, Hu CY, Hentze H, Tan YC et al. SB1518, a novel macrocyclic pyrimidine-based JAK2 inhibitor for the treatment of myeloid and lymphoid malignancies. Leukemia 2011; 25: 1751–1759.
Han L, Qiu P, Zeng Z, Jorgensen JL, Mak DH, Burks JK et al. Single-cell mass cytometry reveals intracellular survival/proliferative signaling in FLT3-ITD-mutated AML stem/progenitor cells. Cytometry 2015; 87: 346–356.
Behbehani GK, Samusik N, Bjornson ZB, Fantl WJ, Medeiros BC, Nolan GP . Mass cytometric functional profiling of acute myeloid leukemia defines cell-cycle and immunophenotypic properties that correlate with known responses to therapy. Cancer Discov 2015; 5: 988–1003.
Sessa C, Shapiro GI, Bhalla KN, Britten C, Jacks KS, Mita M et al. First-in-human phase I dose-escalation study of the HSP90 inhibitor AUY922 in patients with advanced solid tumors. Clin Cancer Res 2013; 19: 3671–3680.
Blanco-Aparicio C, Carnero A . Pim kinases in cancer: diagnostic, prognostic and treatment opportunities. Biochem Pharmacol 2013; 85: 629–643.
Ribeiro D, Melão A, Barata JT . IL-7R-mediated signaling in T-cell acute lymphoblastic leukemia. Adv Biol Regul 2013; 53: 211–222.
Sang L, Roberts JM, Coller HA . Hijacking HES1: how tumors co-opt the anti-differentiation strategies of quiescent cells. Trends Mol Med 2010; 16: 17–26.
Ciceri P, Müller S, O'Mahony A, Fedorov O, Filippakopoulos P, Hunt JP et al. Dual kinase-bromodomain inhibitors for rationally designed polypharmacology. Nat Chem Biol 2014; 10: 305–312.
Lu J, Qian Y, Altieri M, Dong H, Wang J, Raina K et al. Hijacking the E3 ubiquitin ligase cereblon to efficiently target BRD4. Chem Biol 2015; 22: 755–763.
Toure M, Crews CM . Small-molecule PROTACS: new approaches to protein degradation. Agnew Chem Int Ed Engl 2016; 55: 1966–1973.
Liu S, Walker SR, Nelson EA, Cerulli R, Xiang M, Toniolo PA et al. Targeting STAT5 in hematologic malignancies through inhibition of the bromodomain and extra-terminal (BET) bromodomain protein BRD2. Mol Cancer Ther 2014; 13: 1194–1205.
Shih AH, Abdel-Wahab O, Patel JP, Levine RL . The role of mutations in epigenetic regulators in myeloid malignancies. Nat Rev Cancer 2012; 12: 599–612.
Hnisz D, Schuijers J, Lin CY, Weintraub AS, Abraham BJ, Lee TI et al. Convergence of developmental and oncogenic signaling pathways at transcriptional super-enhancers. Mol Cell 2015; 58: 362–370.
Taldone T, Ochiana SO, Patel PD, Chiosis G . Selective targeting of the stress chaperome as a therapeutic strategy. Trends Pharmacol Sci 2014; 35: 592–603.
Zong H, Gozman A, Caldas-Lopes E, Taldone T, Sturgill E, Brennan S et al. A hyperactive signalosome in acute myeloid leukemia drives addiction to a tumor-specific Hsp90 species. Cell Rep 2015; 13: 2159–2173.
Akahane K, Sanda T, Mansour MR, Radimerski T, DeAngelo DJ, Weinstock DM et al. HSP90 inhibition leads to degradation of the TYK2 kinase and apoptotic cell death in T-cell acute lymphoblastic leukemia. Leukemia 2016; 30: 219–228.
Meyer SC, Keller MD, Chiu S, Koppikar P, Guryanova OA, Rapaport F et al. CHZ868, a type II JAK2 inhibitor, reverses type I JAK inhibitor persistence and demonstrates efficacy in myeloproliferative neoplasms. Cancer Cell 2015; 28: 15–28.
Acknowledgements
We thank the Functional Proteomics RPPA Core facility that is supported by the MD Anderson Cancer Center Support Grant 5P30 CA016672-40. The initial heatmaps were developed by the MD Anderson Cancer Center Department of Bioinformatics and Computational Biology, In Silico Solutions, Santeon and SRA International. This work was supported in part by the US National Cancer Institute (NCI; MD Anderson TCGA Genome Data Analysis Center) Grant Numbers CA143883 and CA083639, the Mary K Chapman Foundation and the Michael & Susan Dell Foundation (honoring Lorraine Dell). Additional support was also provided by CPRIT Metabolomics Core Facility Support Award RP120092 (CC), a pilot grant from Alkek Center for Molecular Discovery (CC). This research is supported in part by the MD Anderson Cancer Center Support Grant (P30 CA016672) and the MD Anderson Cancer Center Leukemia SPORE (P50 CA100632).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Dr Pemmaraju serves on the scientific advisory board of Incyte Pharmaceuticals. He has also served as a consultant and received research funding from Novartis Pharmaceuticals. The other authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Leukemia website
Rights and permissions
About this article
Cite this article
Saenz, D., Fiskus, W., Manshouri, T. et al. BET protein bromodomain inhibitor-based combinations are highly active against post-myeloproliferative neoplasm secondary AML cells. Leukemia 31, 678–687 (2017). https://doi.org/10.1038/leu.2016.260
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2016.260
This article is cited by
-
Targeted Therapy for MPNs: Going Beyond JAK Inhibitors
Current Hematologic Malignancy Reports (2023)
-
New Treatments for Myelofibrosis
Current Treatment Options in Oncology (2023)
-
UM171 cooperates with PIM1 inhibitors to restrict HSC expansion markers and suppress leukemia progression
Cell Death Discovery (2022)
-
The BET bromodomain inhibitor ZEN-3365 targets the Hedgehog signaling pathway in acute myeloid leukemia
Annals of Hematology (2021)
-
Novel Targeted Therapeutics in Acute Myeloid Leukemia: an Embarrassment of Riches
Current Hematologic Malignancy Reports (2021)