Abstract
While clinical benefit of the proteasome inhibitor (PI) bortezomib (BTZ) for multiple myeloma (MM) patients remains unchallenged, dose-limiting toxicities and drug resistance limit the long-term utility. The E3 ubiquitin ligase Skp1–Cullin-1–Skp2 (SCFSkp2) promotes proteasomal degradation of the cell cycle inhibitor p27 to enhance tumor growth. Increased SKP2 expression and reduced p27 levels are frequent in human cancers and are associated with therapeutic resistance. SCFSkp2 activity is increased by the Cullin-1-binding protein Commd1 and the Skp2-binding protein Cks1B. Here we observed higher CUL1, COMMD1 and SKP2 mRNA levels in CD138+ cells isolated from BTZ-resistant MM patients. Higher CUL1, COMMD1, SKP2 and CKS1B mRNA levels in patient CD138+ cells correlated with decreased progression-free and overall survival. Genetic knockdown of CUL1, COMMD1 or SKP2 disrupted the SCFSkp2 complex, stabilized p27 and increased the number of annexin-V-positive cells after BTZ treatment. Chemical library screens identified a novel compound, designated DT204, that reduced Skp2 binding to Cullin-1 and Commd1, and synergistically enhanced BTZ-induced apoptosis. DT204 co-treatment with BTZ overcame drug resistance and reduced the in vivo growth of myeloma tumors in murine models with survival benefit. Taken together, the results provide proof of concept for rationally designed drug combinations that incorporate SCFSkp2 inhibitors to treat BTZ resistant disease.
Similar content being viewed by others
Introduction
Clinical success of the proteasome inhibitor (PI) bortezomib (BTZ) (Velcade) established the ubiquitin (Ub)+proteasome system as a key therapeutic target in multiple myeloma (MM).1, 2, 3 While the survival benefit of BTZ has generated new treatment strategies and brought excitement to the community, significant challenges remain. Many patients do not respond to proteasome inhibitor therapy and drug resistance nearly uniformly develops, even in those that initially respond to treatment.4, 5 Moreover, individual patient response to BTZ remains highly variable and the molecular features responsible for the variability in response remain undefined.6, 7, 8, 9
Specificity within the Ub+proteasome system relies upon the selectivity of E3 Ub ligases that maintain proteostasis by targeting individual proteins for proteasomal degradation.10, 11 BTZ blocks the bulk of Ub-dependent protein degradation while drugs that target an individual E3 Ub ligase are expected to destabilize a single protein to confer refined selectivity with reduced adverse toxicities.12, 13 The S-phase kinase associated protein-1 (Skp1) and Cullin-1 bind a multitude of substrate-binding F-box proteins to form multimeric SCF complexes.14, 15, 16 Cell cycle progression is regulated by SCFSkp2, composed of Skp1, Cullin-1 and Skp2, that mediates ubiquitination of the cyclin-dependent kinase (CDK) inhibitor (CKI) p27.17, 18 Progression from G1 to S phase is positively regulated by CDK2 and CDK4, and negatively regulated by p27. SCFSkp2-mediated ubiquitination marks p27 for degradation that permits the CDK-dependent transition from a quiescent to proliferative state. Skp2 binds p27 to facilitate its ubiquitination, and SKP2 expression contributes to increased p27 turnover and enhanced proliferation.19, 20, 21 Cullin-1 scaffolds Skp1 and Skp2 and contributes to proliferation by promoting CKI degradation.22, 23 CUL1 overexpression also promotes proliferation through p27 degradation and high CUL1 expression has been correlated with reduced survival.24, 25, 26, 27 SCF activity is regulated by accessory proteins, for example, Commd1, that promotes SCF ubiquitination activity.28, 29 COMMD1 overexpression is associated with poor outcomes in lymphomas.30 p27 ubiquitination also requires the CDK regulator Cks1 and the Cullin-1-binding protein Rbx1.31
Here publically available databases were used to correlate gene expression in MM patient tumor cells with clinical responses to BTZ. A similar approach recently revealed that nicotinamide phosphoribosyltransferase (Nampt) mRNA expression was higher in BTZ-resistant patient cells and that Nampt represented a viable therapeutic target to overcome BTZ resistance.32 We reveal significantly higher COMMD1 and CUL1 mRNA in patients that did not respond to BTZ. The findings prompted us to investigate the effect of genetic and pharmacologic disruption of the SCFSkp2 complex on BTZ resistance. Using in vitro and in vivo models, we demonstrate that combining a novel SCFSkp2 inhibitor (DT204) with BTZ triggered synergistic anti-myeloma activity and overcame drug resistance.
Materials and methods
Gene expression profile analysis
Cluster version 2.0 was used to analyze data sets GSE9782, GSE2658 and GSE5900.33, 34, 35 Gene expression profiles from tumor cells of patients included in the SUMMIT36 (025), CREST37 phase 2, APEX38 phase 3 trial (039) and HOVON-65/GMMG-HD4 trials35 were analyzed. A two-step filter was used to identify genes differentially regulated in responders vs non-responders. A true statistical test was applied using t-test or analysis of variance to determine if between-condition variability was greater than within-condition variability. Genes identified above with a nominal P-value <0.05 were further refined by applying a fold-change requirement to identify genes with a significant differential. Fold-change was defined as the difference in median expression.
Cell lines and reagents
MM cell lines RPMI8226 and MM1.S (National Cancer Institute, Bethesda, MD, USA) and U266 cells (American Tissue Culture Collection, Manassas, VA, USA) were cultured as described.39, 40 PI-resistant cells were generated as described.40, 41, 42 Hemagglutinin (HA)-tagged Cullin-1 was from Addgene (Cambridge, MA, USA). Bone marrow (BM) aspirates were obtained from patients’ Institutional Review Board approval. Mononuclear cells were removed and malignant plasma cells (PCs) purified by microbead selection (Miltenyi Biotec, San Diego, CA, USA). Antibodies were directed against p27 (Santa Cruz Biotechnology, Dallas, TX, USA), phospho-Thr187 p27, Cks1, Skp1, Skp2, Commd1 and GFP (Abcam, Cambridge, MA, USA) and Cullin-1 and HA (Cell Signaling Technology, Danvers, MA, USA). Bands were visualized using LiCoR (Littleton, CO, USA) IRDye goat anti-mouse or donkey anti-rabbit antibodies.
Cell viability
Cells viability was determined using the XTT (2,3-Bis-(2-Methoxy-4-Nitro-5-Sulfophenyl)-2H-Tetrazolium-5-Carboxanilide) assay. Relative viability was calculated based on that of untreated cells. Cells were stained with annexin-V fluorescein isothiocyanate (Thermo-Fisher, Waltham, MA, USA) and analyzed on a BD-LSR-Fortessa flow cytometer (BD Biosciences, San Jose, CA, USA) to quantitate apoptotic cells.
In vivo murine model of MM
To evaluate the effect of DT204 in vivo, 5-week-old female athymic NCr (nu/nu) mice were injected in the tail vein with 2 × 106 MM1.S-luciferase-expressing cells according to the Institutional Review Board-approved protocols. Mice were randomized to four groups (7 or 8/group) and received intravenous injection of vehicle (phosphate-buffered saline in 10% dimethylsulfoxide); BTZ (0.5 mg/kg), DT204 (10 mg/kg) or DT204+BTZ administered on days 1, 3, 5, 7 and 9. Mice were imaged by bioluminescence imaging at days 0 and 14. Animal body weights were measured every third day from start of treatment. Treatment effect on overall survival (OS) was determined using Kaplan–Meier curves.
Mice were killed when tumors reached 2 cm3, became ulcerated or elicited complications that limited mobility and feeding.
Statistical analysis
In vitro assays were performed in triplicate. Statistical significance of differences was determined using the Student’s t- and analysis of variance tests with a minimal level of significance of P<0.05. In vivo statistical tests were performed using the two-tailed Student’s t-test. Median OS was determined using the Kaplan–Meier method with 95% confidence intervals.43 Prism Version 6.0 software (GraphPad, San Diego, CA, USA) was used for statistical analyses.
Drug synergy
The Compusyn software and program was used to determine the effect of drug combinations and general dose effects and is based on the median-effect principle and the Chou–Talalay Combination Index–Isobologram Theorem.44 The Chou–Talalay method for drug combination is based on the median-effect equation, derived from the mass-action law principle, which provides the common link between single entity and multiple entities, and first-order and higher-order dynamics. The resulting combination index (CI) theorem of Chou–Talalay offers quantitative definition for additive effect (CI=1), synergism (CI<1) and antagonism (CI>1) in drug combinations.
Results
Expression of SCFSkp2 components correlates with MM patient survival
The goal of the present study was to identify genes within the Ub+proteasome system that were upregulated in tumor cells from MM patients that did not respond to BTZ as these genes represent potentially actionable therapeutic targets. We analyzed data sets from patients enrolled in pharmacogenomic studies performed to compare the efficacy of BTZ with other anti-myeloma agents. Prospectively collected BM-derived PCs were analyzed to identify candidates that correlated with treatment outcome. To determine whether expression of the SCFSkp2 components offered prognostic benefit, patients that received BTZ were stratified based upon gene expression and progression-free survival (PFS) or OS. CUL1, COMMD1, SKP2 and CKS1B expression above the median value was associated with significantly decreased PFS and OS (Figure 1a). SKP2 and CKS1B expression also correlated with reduced OS in patients treated with BTZ in the HOVON-65/GMMG-HD4 trial (Figure 1b). In trial 039, patients received treatment with either BTZ or dexamethasone. As CUL1, SKP2, COMMD1 and CKS1B expression negatively correlated with PFS and OS, we investigated whether the expression of these genes also correlated with treatment response (Figure 1c). The results indicated that the expression of COMMD1 and CUL1 was negatively correlated with the response to BTZ. However, the expression of these same genes did not correlate with the response to dexamethasone (Figure 1c). A heat map generated from DNA microarray data also indicated that the expression of CUL, COMMD1 and SKP2 was significantly greater in tumor cells from BTZ-non-responders compared with BTZ-responders (Supplementary Figure 1A). Genes upregulated in BTZ-responders were rank-ordered to indicate that COMMD1, CUL1 and SKP2 were among the most highly upregulated (Supplementary Figure 1B). COMMD1, CUL1 and SKP2 expression was then compared in PCs from healthy individuals and monoclonal gammopathy of unknown significance patients with those of smoldering MM and MM patients (Supplementary Figure 1C). COMMD1, CUL1 and SKP2 expression was greater in smoldering MM and MM patients than in healthy individuals and monoclonal gammopathy of unknown significance patients.
Correlation of the expression of SCFSkp2 components with survival and response to BTZ. (a) Kaplan–Meier PFS curves from MM patients in APEX trial (039) after BTZ treatment (n=163) according to gene expression profile arrays generated at Millenium Pharmaceuticals (Cambridge, MA, USA GSE9782). Probes used were CUL1-207614, SKP2-210567, COMMD1-226024 and CKS1B-201897. PFS was plotted using median values determined for each probe based on data set GSE9782 obtained from analysis of patient samples. Kaplan–Meier OS curves of patients treated with BTZ according to CUL1, SKP2, COMMD1 and CKS1B expression above or below median values (GSE9782) are also shown. (b) Kaplan–Meier OS curves for the expression of either SKP2 or CKS1B based on their expression in patients enrolled in the HOVON65/GMMG-HD4 study (GSE19784). Expression of the following probe sets was evaluated: CKS1B (201897); COMMD1 (226024); CUL1 (207614 and 238509); RBX1 (218117); SKP1 (200711 and 200719); and SKP2 (210567 and 203625). Differences in expression in the response categories were assessed using Kruskal–Wallis analysis to distinguish four categories: complete response/near complete response (CR/nCR); very good partial response (VGPR); partial response (PR); and minimal response (MR) or worse. The number of patients per category for the BTZ arm was 17 (CR/nCR), 55 (VGPR), 63 (PR) and 11 (MR or worse), and for the control arm was 4 (CR/nCR), 16 (VGPR), 66 (PR) and 43 (MR or worse). Survival analysis was performed as described using PFS censored for allogeneic transplant and OS.39 Survival analysis was performed with 169 cases in the BTZ arm and 158 cases in the control arm. (c) Expression of COMMD1, CUL1, SKP2 and CKS1B in tumor cells from patients enrolled in trial 039 that had been treated with BTZ (n=163) or dexamethasone (DEX; n=70). Response to therapy was defined in trial 039 and HOVON-65/GMMG-HD4 using criteria established by the European Group for BMT: http://www.ncbi.nlm.nih.gov/pubmed/9753033?access_num=9753033&link_type=MED&sso-checked=true&dopt=Abstract. Responders were patients that achieved a complete remission (CR), very good partial remission (VGPR), partial remission (PR) or minor remission (MR), and non-responders were those patients that achieved progressive disease (PD), stable disease or minor remission. PD required a 25% increase in paraprotein, whereas MR, PR and CR required at least a 25%, 50% or 100% decrease, respectively. Baseline correction of the original data set was performed to identify individual genes differentially expressed in BTZ-non-responders (NRs) relative to BTZ-responders (Rs). Analysis criteria required at least a fivefold log difference in expression between the NRs and Rs and P-value <0.05. COMMD1 expression difference in BTZ-NRs vs BTZ-Rs was log (fold-change)=151 (P=1.3E-2) and in the DEX-NRs vs DEX-Rs was log (fold-change)=77 (P=4.4E-1). For CUL1, expression difference in BTZ-NRs vs BTZ-Rs was log (fold-change)=32 (P=2.8E-4) and in DEX-NRs vs DEX-Rs the change was log (fold-change)=−21 (P=1.1E-1). Probes used were SKP1-200711, 200718, 200719, 2007974; SKP2-203625, 203626, 210567; CULLIN-1-207614; COMMD1-226024; RBX1-218117; and CKS1B-201897.
CUL1, COMMD1 or SKP2 knockdown sensitizes cells to proteasome inhibitors
To demonstrate a role for CUL1, COMMD1 and SKP2 in BTZ-induced cytotoxicity, parental (drug-sensitive) MM cells were transfected with either control (scrambled) short hairpin RNA (shRNA) or shRNA directed against CUL1, COMMD1 or SKP2. Target knockdown was confirmed by quantitative reverse transcription–PCR and western blot (Supplementary Figures 2A and B). MM cells were then treated with different PIs and the number of annexin-V-positive cells was quantitated (Figure 2). The results indicated that knockdown of CUL1, COMMD1 or SKP2 increased the number of annexin-V-positive cells after PI treatment. For example, after knockdown of CUL1, COMMD1 or SKP2, BTZ treatment (10 nM) yielded 38, 40 and 42% annexin-V-positive cells compared with 17% in controls. In contrast, treatment with lenalidomide did not significantly increase the number of annexin-V-positive cells in cells after the knockdown of CUL1, COMMD1 or SKP2. A similar effect of CUL1, COMMD1 and SKP2 knockdown on the cytotoxic effect of PIs was also observed with U266 cells (Supplementary Figure 2C).
CUL1 knockdown sensitizes BTZ-resistant cells to proteasome inhibition. Drug-sensitive or -resistant cells were transfected with control shRNA or shRNA to specifically knockdown CUL1, COMMD1 or SKP2. Transfectants were then treated as indicated and the number of annexin-positive cells determined in triplicate.
Knockdown of CUL1, COMMD1 and SKP2 overcomes resistance to PIs
To further investigate the role of CUL1, COMMD1 and SKP2 in the therapeutic response to PIs, RPMI and U266 cells were exposed to increased concentrations of the PIs BTZ, carfilzomib or ixazomib to generate models of PI resistance. Gene sequencing demonstrated that the proteasome catalytic gene PSMB5 was not mutated in the RPMI8226 and U266 cells generated with drug-resistance to BTZ, carfilzomib or ixazomib (Supplementary Figure 7). Quantitative reverse transcription–PCR indicated that CUL1, COMMD1 and SKP2 expression levels were greater in the RPMI8226 and U266 PI-resistant cells compared with the parental cells (Figure 3a and Supplementary Table 1). Western blots indicated that the protein level of Cullin-1, Commd1 and Skp2 was increased in the PI-resistant RPMI8226 and U266 cells (Figure 3b). The PI-resistant RPMI8226 and U266 cells also exhibited increased proteasome catalytic activity, consistent with prior related studies (Supplementary Table 2). This is consistent with previous reports supporting the notion that BTZ-resistant cells contain more proteasomes and/or more proteasome activity.45, 46, 47 Also, DNA sequencing indicated that PSMB5, the gene that encodes the proteasome subunit responsible for chymotrypsin-like activity, was not mutated in the RPMI8226 cells resistant to BTZ, ixazomib or carfilzomib. Sequencing also revealed that PSMB5 was not mutated in drug-sensitive or -resistant U266 cells. PI-resistant cells were then transfected with shRNA to reduce CUL1, COMMD1 or SKP2 levels that was confirmed by quantitative reverse transcription–PCR and western blot (Supplementary Figure 3). CUL1, COMMD1 or SKP2 knockdown increased the sensitivity of PI-resistant RPMI8826 (Figure 3c) and U266 (Supplementary Figure 3) cells to BTZ, carfilzomib or ixazomib.
Relative expression of SCFSkp2 components in BTZ-sensitive or -resistant cells. (a) Quantitative reverse transcription (qRT–PCR) of SKP2, COMMD1 and CUL1 expression in RPMI8226 BTZ-sensitive (S) or -resistant (R) cells. Coding sequences (5′–3′) were based on the NCBI AceView BLAST system. qRT–PCR primer sequences (5′–3′) were as follows: SKP2 forward-CAGGCCTAAGCTAAATCGAGAG, SKP2 reverse-CTGGCAATGGTGGTGAAATG; CUL1 forward-CAGAGGAGGCAGAACTAGAAGA, CUL1 reverse-CGTGTTGTACTGAAGCAGGATAG; and COMMD1 forward AGCCAGCTATATCCAGAGGT, COMMD1 reverse CATGAGGCTCTCACGGATT. Total RNA was isolated using the RNeasy mini kit (Qiagen, Germantown, MD, USA). cDNA synthesis was performed using the SuperScript III first-Strand Synthesis System. qRT–PCR was performed using iTaq Universal Sybr Green Supermix (Bio-Rad, Hercules, CA, USA) in the CFX96 Real-Time PCR detection system (Bio-Rad). Polymerase activation and DNA denaturation was performed at 95 °C for 30 s followed by 40 cycles of denaturation at 95 °C for 5 s, annealing at 50 °C for 30 s, extension at 60 °C for 30 s and the melting curve analyzed at 65–95 °C. Calculations were performed using the 2−ΔΔCt method, where ΔCt is the Ct value of GADPH subtracted from the Ct value of the gene of interest. ΔΔCt is the ΔCt value of the control subtracted from the ΔCt value of the gene of interest. Error bars were determined based on triplicate SKP1 expression that was not significantly different in parental vs resistant samples. (b) Western blot of Cullin-1, Commd1 and Skp2 in parental and PI-resistant cells. Lysates were separated by SDS–polyacrylamide gel electrophoresis, transferred to nitrocellulose membranes and probed using indicated antibodies. (c) Effect of shRNA knockdown of CUL1, COMMD1 and SKP2 on generation of annexin-V-positive cells. shRNA in the pLKO.1-TCR cloning vector were transfected into 293T packaging cells with each plasmid, packaging envelope and lipofectamine-2000. Viral supernatants were concentrated and resuspended in phosphate-buffered saline at 1 × 109 infectious units per ml, and used to transduce cells then selected in puromycin. Cells were treated as indicated and annexin-positive cells then quantitated in triplicate.
Physical association of Cul1 and Commd1
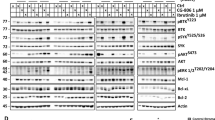
RPMI8226 and U266 cells were transfected with control shRNA or shRNA directed against COMMD1 or CUL1. Transfectants were probed by western blot to demonstrate that COMMD1 or CUL1 knockdown increased p27 levels (Figure 4a). Parental and PI-resistant cells were then transfected with a plasmid that expressed HA-tagged Cullin-1 and treated as indicated. Lysates were prepared, immunoprecipitated using an HA-specific antibody and probed by western blot. Results indicated that Commd1 was associated with HA-Cullin-1 in RPMI8226 and U266 cells, and that the association was increased after BTZ treatment in both parental and PI-resistant cells (Figure 4b).
Commd1 and Cullin1 regulation of p27. (a) Western blot of p27 from lysates after transfection with scrambled, COMMD1 or CUL1 shRNA. Cell lysates were prepared as previously described, separated by SDS–polyacrylamide gel electrophoresis and probed with indicated antibodies.40 (b) Association of Commd1 with HA-tagged Cullin-1. Western blot of HA immunoprecipitates from PI-sensitive or -resistant RPMI826 or U266 cells transfected with a plasmid that expressed HA-CUL-1 after PI treatment. Lysates were immunoprecipitated with IgG as controls (lanes 1 and 2).
Identification of a novel SCFSkp2 inhibitor
SCFSkp2 inhibitors offer an attractive approach to pharmacologically target the Ub+proteasome system with more specificity than currently available with PIs.42, 48, 49 On the basis of structural features of Skp2, as well as those of known Skp2 inhibitors, we performed virtual and computational screening to identify a panel of novel compounds as potential SCFSkp2 inhibitors. RPMI8226 cells were transfected with pBABE-MN-IRES-E-GFP-p27, a plasmid that expressed green fluorescent protein-tagged p27 to assess the effect of these compounds on p27 stability (Supplementary Figure 4A). Compounds were incubated with the transfected cells to identify those that increased green fluorescence as a read-out of p27 stability (Figure 5a). DT204 significantly increased cellular fluorescence to a level greater than that observed with known Skp2 inhibitors. Western blotting indicated that DT204 also increased p27 levels in cell lysates (Supplementary Figures 4B and C). In contrast to BTZ or carfilzomib, DT204 treatment did not increase the level of p53. DT204 co-treatment with BTZ enhanced the generation of annexin-V-positive cells compared with BTZ alone in a number of different MM cell lines (Figure 5b). The Chou–Talaly theorem and Compusyn software were used to detect potential synergy between DT204 and BTZ. The CI values for DT204 combined with BTZ at 1 nM were 0.7, 0.8 and 0.7 for RPMI8226, U266 and MM1.S, respectively, to indicate formal synergy when tested within the three different MM cell lines.
DT204 effect on SCFSkp2 and p27 levels. (a) Compounds were added at 10 or 50 μM for 24 h and green fluorescence in cells transfected with the green fluorescent protein-p27-expressing vector was measured. (b) DT204 co-treatment with BTZ increases the amount of annexin-V-positive cells. Annexin-V-positive cells were quantitated by flow cytometry. BTZ treatment alone at indicated concentrations produced a negligible amount of annexin-V-positive cells.
DT204 co-treatment with BTZ overcomes drug resistance
DT204 co-treatment induced apoptosis in cells resistant to the proteasome inhibitors BTZ, carfilzomib or ixazomib (Figure 6). CI values for the combination of DT204 and BTZ to treat BTZ-R, CFZ-R an IXZ-R cells were <1, consistent with formal synergy. We observed cross-sensitization, that is, DT204 sensitized BTZ-R cells to not only BTZ but also CFZ and IXZ.
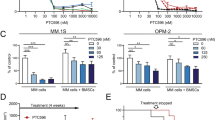
DT204 co-treatment with BTZ reduces MM patient PC growth and colony formation in the presence of BMSCs
CD138+ PCs were isolated from MM patient BM and treated with DT204, BTZ or both (Figure 7a). DT204 co-treatment with BTZ synergistically reduced the viability of MM patient CD138+ PCs. Patient PCs were then cultured alone or co-cultured with BMSCs and treated with DT204, BTZ or both. The number of colonies formed by patient PCs was then measured to indicate that DT204 co-treatment with BTZ reduced the number of colonies formed after co-culture with BMSCs (Figure 7b).
DT204 effect on MM patient PC viability and colony formation. (a) DT204 effect alone or combined with BTZ was determined using patient CD138+ cells. (b) Effect of BTZ and DT204 on colony formation from patient CD138+ cells cultured alone or with stromal cells. CD138+ cells were labeled with carboxyfluorescein succinimidyl ester, co-cultured with HS.5 cells, treated as indicated and green colonies counted.
DT204+BTZ triggers synergistic inhibition of MM xenograft growth in vivo
We determined the in vivo efficacy of DT204 in combination with BTZ using bioluminescent MM cells injected through the tail vein into murine models. Mice treated with DT204, BTZ or DT204+BTZ showed a statistically significant delay in tumor growth compared with vehicle-treated mice (Figure 8a). DT204 was well-tolerated and did not provoke overt dermatologic, musculoskeletal, neurologic or body weight changes (Figure 8b). A statistically significant prolongation in median OS was observed in the DT204+BTZ-treated group (34 days) compared with vehicle-treated group (12 days), or in the group treated with DT204 BTZ alone (24 and 26 days, Figure 8c). Western blot of tumor collected from mice following treatment with DT204 or DT204+BTZ showed a greater accumulation of p27 compared with the other treatment groups (Figure 4d). The results suggest that synergistic activity observed at the cellular level with DT204 in combination with BTZ translates to in vivo efficacy.
DT204+BTZ triggers synergistic inhibition of MM xenograft growth in vivo. (a) Representative bioluminescence imaging of mice from each treatment group at day 18 after treatment initiation. (b) Animal body weight after treatment with vehicle, DT204, BTZ or DT204+BTZ combination. (c) Kaplan–Meier survival curve comparison of nude mice after subcutaneous injection of myeloma cells treated with vehicle, DT204, BTZ or DT204+BTZ. (d) Western blot of p27 from tumor tissue of mice treated with vehicle, BTZ, DT204 or both.
Discussion
There remains an urgent and unmet need to define the molecular mechanisms of BTZ resistance in order to enhance the use of existing treatments and design more effective single-agent and combination therapies. In the current study, we used a biologically supervised approach that identified SCFSkp2 components as molecular classifiers expressed at greater levels in tumors from MM patients that did not respond to BTZ. Expression of COMMD1, CUL1, SKP2 and CKS1B in tumors from relapsed/refractory patients was also negatively correlated with PFS and OS. The same genes were not significantly upregulated in patients that failed to respond to other anti-myeloma therapies and may therefore serve as classifiers to stratify BTZ-based therapy.39 Cell-based assays demonstrated that shRNA-mediated knockdown of COMMD1, CUL1 or SKP2 enhanced the sensitivity of MM cells to BTZ and synergistically enhanced the effect of BTZ. Furthermore, SCFSkp2 has previously been associated with tumor progression and drug resistance,21, 25, 26, 27 to suggest that SCFSkp2 inhibitors may restore drug sensitivity to overcome BTZ resistance.
We identified a novel compound that inhibited SCFSkp2-mediated degradation of p27 and enhanced the anti-myeloma effect of BTZ in MM cell lines and patient samples. DT204 stabilized p27, reduced myeloma viability, synergistically enhanced the anti-myeloma effect of BTZ and prevented Skp2 incorporation into the SCFSkp2 complex. DT204 was not toxic to inactive or phytohemagglutinin-activated peripheral blood mononuclear cells and promoted cell cycle arrest (Supplementary Figure 4). Genetic knockout of SKP2 similarly promoted cell cycle arrest and DT204 phenocopied the effect of SKP2 deficiency on cell cycle arrest (Supplementary Figure 5). DT204 also displayed p27-independent effects as observed by the effect on the viability of cells transfected with a phospho-deficient (T187A) p27 mutant (Supplementary Figure 6). Finally, we observed that DT204 enhanced the effect of BTZ in a xenograft model of myeloma. The marked antitumor activity of this combination, coupled with the lack of its effect on peripheral blood mononuclear cells, suggests a favorable therapeutic index.
The present studies link individual components of the SCFSkp2 complex to BTZ resistance and form the genetic basis to prospectively define those patients that should benefit from BTZ therapy. The findings provide the rationale for the advancement of genomic biomarker-based evaluation of patients for whom BTZ is being considered. These studies also provide proof of concept to target SCFSkp2 as a strategy to overcome therapeutic resistance and provide the rationale for the development of novel drug combinations to enhance the efficacy of BTZ-based therapies in MM. In summary, we demonstrate that the SCFSkp2-p27 axis represents a major determinant in the ability of BTZ to induce apoptosis.
References
Palumbo A, Anderson K . Multiple myeloma. N Engl J Med 2011; 364: 1046–1060.
Richardson PG, Mitsiades CS, Hideshima T, Anderson KC . Novel biological therapies for the treatment of multiple myeloma. Best Pract Res Clin Haematol 2005; 18: 619–634.
Richardson PG, Schlossman RL, Alsina M, Weber DM, Coutre SE, Gasparetto C et al. PANORAMA 2: panobinostat in combination with BTZ and dexamethasone in patients with relapsed and BTZ-refractory myeloma. Blood 2013; 122: 2331–2337.
Chauhan D, Tian Z, Nicholson B, Kumar KG, Zhou B, Carrasco R et al. A small molecule inhibitor of ubiquitin-specific protease-7 induces apoptosis in multiple myeloma cells and overcomes BTZ resistance. Cancer Cell 2012; 22: 345–358.
Maiso P, Huynh D, Moschetta M, Sacco A, Aljawai Y, Mishima Y et al. Metabolic signature identifies novel targets for drug resistance in multiple myeloma. Cancer Res 2015; 75: 2071–2082.
Bolli N, Avet-Loiseau H, Wedge DC, Van Loo P, Alexandrov LB, Martincorena I et al. Heterogeneity of genomic evolution and mutational profiles in multiple myeloma. Nat Commun 2014; 5: 2997.
Morgan GJ, Walker BA, Davies FE . The genetic architecture of multiple myeloma. Nat Rev Cancer 2012; 12: 335–348.
Walker BA, Wardell CP, Melchor L, Hulkki S, Potter NE, Johnson DC et al. Intraclonal heterogeneity and distinct molecular mechanisms characterize the development of t(4,14) and t(11;14) myeloma. Blood 2012; 120: 1077–1086.
Lohr JG, Stojanov P, Carter SL, Cruz-Gordillo P, Lawrence MS, Auclair D et al. Widespread genetic heterogeneity in multiple myeloma: implications for targeted therapy. Cancer Cell 2014; 25: 91–101.
Deshaies RJ, Joazeiro CAP . RING domain E3 ubiquitin ligases. Annu Rev Biochem 2009; 78: 399–434.
Emanuele MJ, Elia AEH, Xu Q, Thoma CR, Izhar L, Leng Y et al. Global identification of modular cullin-RING ligase substrates. Cell 2011; 147: 459–474.
Orlowski R . Why proteasome inhibitors cannot ERADicate multiple myeloma. Cancer Cell 2013; 24: 275–277.
Leung-Hagesteijn C, Erdmann N, Cheung G, Keats JJ, Stewart AK, Reece DE . Xbp1s-negative tumor B cells and pre-plasmablasts mediate therapeutic proteasome inhibitor resistance in multiple myeloma. Cancer Cell 2013; 24: 289–304.
Nakayama KI, Nakayama K . Ubiquitin-ligases: cell-cycle control and cancer. Nat Rev Cancer 2009; 6: 269–281.
Jia L, Sun Y . SCF E3 ubiquitin ligases as anticancer targets. Curr Cancer Drug Targets 2011; 11: 347–356.
Bennett E, Rush J, Gygi SP, Harper JW . Dynamics of Cullin-Ring ubiquitin ligase network revealed by systematic quantitative proteomics. Cell 2010; 143: 951–965.
Bornstein G, Bloom J, Sitry-Shevah D, Nakayama K, Pagano M, Hershko A . Role of the SCFSkp2 ubiquitin ligase in the degradation of p21Cip1 in S phase. J Biol Chem 2003; 278: 25752–25757.
Yu ZK, Gervais JL, Zhang H . Human CUL-1 associates with the SKP1/SKP2 complex and regulates p21(CIP1/WAF1) and cyclin D proteins. Proc Natl Acad Sci USA 1998; 95: 11324–11329.
Skowyra D, Craig KL, Tyers M, Elledge SJ, Harper JW . F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell 2007; 91: 209–219.
Frescas D, Pagano M . Deregulated proteolysis by the F-box proteins SKP2 and beta-TrCP: tipping the scales of cancer. Nat Rev Cancer 2008; 8: 438–449.
Gstaiger M, Jordan R, Lim M, Catzavelos C, Mestan J, Slingerland J et al. Skp2 is oncogenic and overexpressed in human cancers. Proc Natl Acad Sci USA 2001; 98: 5043–5048.
Chan CH, Li CF, Yang WL, Gao Y, Lee SW, Feng Z et al. The Skp2-SCF E3 ligase regulates Akt ubiquitination, glycolysis, herceptin sensitivity, and tumorigenesis. Cell 2012; 149: 1098–1111.
Kipreos ET, Lander LE, Wing JP, He WW, Hedgecock EM . Cul-1 is required for cell cycle exit in C. elegans and identifies a novel gene family. Cell 1996; 85: 829–839.
Wang Y, Penfold S, Tang X, Hattori N, Riley P, Harper JW et al. Deletion of the Cul1 gene in mice causes arrest in early embryogenesis and accumulation of cyclin E. Curr Biol 1999; 9: 1191–1194.
Chen G, Li G . Increased CUL1 expression promotes melanoma cell proliferation through regulating p27 expression. Int J Oncol 2010; 37: 1339–1344.
Bai J, Zhou Y, Chen G, Zeng J, Ding J, Tan Y et al. Overexpression of Cullin1 is associated with poor prognosis of patients with gastric cancer. Hum Pathol 2011; 42: 375–383.
Min KW, Kim DH, Do SI, Sohn JH, Chae SW, Pyo J et al. Diagnostic and prognostic relevance of Cullin1 expression in invasive ductal carcinoma of the breast. J Clin Pathol 2012; 65: 896–901.
Wu S1, Zhu W, Nhan T, Toth JI, Petroski MD, Wolf DA . CAND1 controls in vivo dynamics of the cullin 1-RING ubiquitin ligase repertoire. Nat Commun 2013; 4: 1642.
Mao X, Gluck N, Li H, Chen B, Wallis M, Maine GN et al. Copper metabolism MURR1 domain containing 1 (COMMD1) regulates Cullin-RING ligases by preventing Cullin-associated NEDD8-dissociated (CAND1) binding. J Biol Chem 2011; 286: 32355–32365.
Taskinen M, Louhimo R, Koivula S, Chen P, Rantanen V, Holte H . Deregulation of COMMD1 is associated with poor prognosis in diffuse large B-cell lymphoma. Plos One 2014; 9: e91031.
Zhan F, Colla S, Wu X, Chen B, Stewart JP, Kuehl WM et al. CKS1B, overexpressed in aggressive disease, regulates multiple myeloma growth and survival through SKP2- and p27Kip1-dependent and -independent mechanisms. Blood 2007; 109: 4995–5001.
Cagnetta A, Cea M, Calimeri T, Acharya C, Fulciniti M, Tai YT et al. Intracellular NAD(+) depletion enhances BTZ-induced anti-myeloma activity. Blood 2013; 122: 1243–1255.
Mulligan G, Mitsiades C, Bryant B, Zhan F, Chng WJ, Roels S et al. Gene expression profiling and correlation with outcome in clinical trials of the proteasome inhibitor BTZ. Blood 2007; 109: 3177–3188.
Zhan F, Hardin J, Kordsmeier B, Bumm K, Zheng M, Tian E et al. Global gene expression profiling of multiple myeloma, monoclonal gammopathy of undetermined significance and normal bone marrow plasma cells. Blood 2002; 99: 1745–1757.
Broyl A, Hose D, Lokhorst H, de Knegt Y, Peeters J, Jauch A et al. Gene expression profiling for molecular classification of multiple myeloma in newly diagnosed patients. Blood 2010; 116: 2543–2553.
Richardson PG, Barlogie B, Berenson J, Singhal S, Jagannath S, Irwin D et al. A phase 2 study of BTZ in relapsed, refractory myeloma. N Engl J Med 2003; 348: 2609–2617.
Jagannath S, Barlogie B, Berenson J, Siegel D, Irwin D, Richardson PG et al. A phase 2 study of two doses of BTZ in relapsed or refractory myeloma. Br J Haematol 2004; 127: 165–172.
Richardson PG, Sonneveld P, Schuster M et al. Extended follow-up of a phase 3 trial in relapsed multiple myeloma: final time-to-event results of the APEX trial. Blood 2007; 110: 3557–3560.
Sonneveld P, Schmidt-Wolf IG, van der Holt B, El Jarari L, Bertsch U, Salwender H et al. Bortezomib induction and maintenance treatment in patients with newly diagnosed multiple myeloma: results of the randomized phase III HOVON-65/GMMG-HD4 trial. J Clin Oncol 2012; 30: 2946–2955.
Jagannathan S, Vad N, Vallabhapurapu Su, Anderson KC, Driscoll JJ . MiR-29b replacement inhibits proteasomes and disrupts aggresome+ autophagosome formation to enhance the anti-myeloma benefit of BTZ. Leukemia 2014; 29: 727–738.
Jagannathan S, Abdel Malek M, Malek E, Vad N, Anderson KC, Driscoll JJ . Pharmacologic screens reveal metformin that suppresses GRP78-dependent autophagy to enhance the anti-myeloma effect of BTZ. Leukemia 2015; 29: 2184–2191.
Wu L, Grigoryan AV, Li Y, Hao B, Pagano M, Cardozo TJ . Specific small molecule inhibitors of Skp2-mediated p27 degradation. Chem Biol 2012; 19: 1515–1524.
Kaplan EL, Meier P . Nonparametric estimation from incomplete observations. J Amer Statist Assn 1958; 53: 457–481.
Chou TC . Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res 2010; 70: 440–446.
Suzuki E, Demo S, Deu E, Keats J, Arastu-Kapur S, Bergsagel PL et al. Molecular mechanisms of BTZ resistant adenocarcinoma cells. Plos One 2011; 6: e27996.
Perez-Galan P, Mora-Jensen H, Weniger MA, Shaffer AL 3rd, Rizzatti EG, Chapman CM et al. Bortezomib resistance in mantle cell lymphoma is associated with plasmacytic differentiation. Blood 2011; 117: 542–552.
Codony-Servat J, Tapia MA, Bosch M, Oliva C, Domingo-Domenech J, Mellado B et al. Differential cellular and molecular effects of BTZ, a proteasome inhibitor, in human breast cancer cells. Mol Cancer Ther 2006; 5: 665–675.
Chan CH, Morrow JK, Li CF, Gao Y, Jin G, Moten A et al. Pharmacological inactivation of Skp2 SCF ubiquitin ligase restricts cancer stem cell traits and cancer progression. Cell 2013; 154: 556–568.
Chen Q, Xie W, Kuhn DJ, Voorhees PM, Lopez-Girona A, Mendy D et al. Targeting the p27 E3 ligase SCF(Skp2) results in p27- and Skp2-mediated cell-cycle arrest and activation of autophagy. Blood 2008; 111: 4690–4699.
Acknowledgements
JD is recipient of the UC Hematology and Oncology Translational Studies Award and a mentor in the Egyptian–US Joint Supervision Program. MAY is recipient of the Egyptian Cultural and Educational Bureau Award in the Egyptian–US Joint Supervision Program. SJ is recipient of the American Society of Hematology Wallace H Coulter Award.
Author contributions
Agree with the manuscript’s results and conclusions: EM, SJ, MAY, NV, RK, AJ, AB, PS, MD, FC, KCA and JD; designed the experiments: EM, MAY, SJ and JD; analyzed the data: EM, SJ, MAY, NV, RK, AJ, AB, PS, MD, FC, KCA and JD; collected data/did experiments for the study: EM, SJ, MAY, NV, RK, AJ, AB, MD, FC and JD; contributed to writing of the paper: EM, MAY, NV, MD, KCA and JD; supervised experiments: JD.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Leukemia website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Malek, E., Abdel-Malek, M., Jagannathan, S. et al. Pharmacogenomics and chemical library screens reveal a novel SCFSKP2 inhibitor that overcomes Bortezomib resistance in multiple myeloma. Leukemia 31, 645–653 (2017). https://doi.org/10.1038/leu.2016.258
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2016.258
This article is cited by
-
The cross talk of ubiquitination and chemotherapy tolerance in colorectal cancer
Journal of Cancer Research and Clinical Oncology (2024)
-
Skp2-mediated MLKL degradation confers cisplatin-resistant in non-small cell lung cancer cells
Communications Biology (2023)
-
E3 ubiquitin ligases in cancer stem cells: key regulators of cancer hallmarks and novel therapeutic opportunities
Cellular Oncology (2023)
-
Flavokawain B targets protein neddylation for enhancing the anti-prostate cancer effect of Bortezomib via Skp2 degradation
Cell Communication and Signaling (2019)
-
Binimetinib, a novel MEK1/2 inhibitor, exerts anti-leukemic effects under inactive status of PI3Kinase/Akt pathway
International Journal of Hematology (2019)