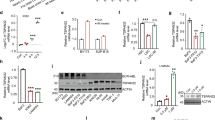
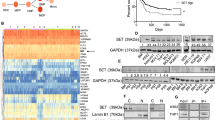
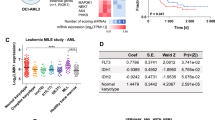
Abstract
The t(8;21) rearrangement, which creates the AML1-ETO fusion protein, represents the most common chromosomal translocation in acute myeloid leukemia (AML). Clinical data suggest that CBL mutations are a frequent event in t(8;21) AML, but the role of CBL in AML1-ETO-induced leukemia has not been investigated. In this study, we demonstrate that CBL mutations collaborate with AML1-ETO to expand human CD34+ cells both in vitro and in a xenograft model. CBL depletion by shRNA also promotes the growth of AML1-ETO cells, demonstrating the inhibitory function of endogenous CBL in t(8;21) AML. Mechanistically, loss of CBL function confers hyper-responsiveness to thrombopoietin and enhances STAT5/AKT/ERK/Src signaling in AML1-ETO cells. Interestingly, we found the protein tyrosine phosphatase UBASH3B/Sts-1, which is known to inhibit CBL function, is upregulated by AML1-ETO through transcriptional and miR-9-mediated regulation. UBASH3B/Sts-1 depletion induces an aberrant pattern of CBL phosphorylation and impairs proliferation in AML1-ETO cells. The growth inhibition caused by UBASH3B/Sts-1 depletion can be rescued by ectopic expression of CBL mutants, suggesting that UBASH3B/Sts-1 supports the growth of AML1-ETO cells partly through modulation of CBL function. Our study reveals a role of CBL in restricting myeloid proliferation of human AML1-ETO-induced leukemia, and identifies UBASH3B/Sts-1 as a potential target for pharmaceutical intervention.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Goyama S, Mulloy JC . Molecular pathogenesis of core binding factor leukemia: current knowledge and future prospects. Int J Hematol 2011; 94: 126–133.
Link KA, Chou FS, Mulloy JC . Core binding factor at the crossroads: determining the fate of the HSC. J Cell Physiol 2010; 222: 50–56.
Cunningham L, Finckbeiner S, Hyde RK, Southall N, Marugan J, Yedavalli VR et al. Identification of benzodiazepine Ro5-3335 as an inhibitor of CBF leukemia through quantitative high throughput screen against RUNX1-CBFbeta interaction. Proc Natl Acad Sci USA 2012; 109: 14592–14597.
Illendula A, Pulikkan JA, Zong H, Grembecka J, Xue L, Sen S et al. Chemical biology. A small-molecule inhibitor of the aberrant transcription factor CBFbeta-SMMHC delays leukemia in mice. Science 2015; 347: 779–784.
Aranaz P, Migueliz I, Hurtado C, Erquiaga I, Larrayoz MJ, Calasanz MJ et al. CBL RING finger deletions are common in core-binding factor acute myeloid leukemias. Leuk Lymphoma 2012; 54: 428–431, 17.
Haferlach C, Dicker F, Kohlmann A, Schindela S, Weiss T, Kern W et al. AML with CBFB-MYH11 rearrangement demonstrate RAS pathway alterations in 92% of all cases including a high frequency of NF1 deletions. Leukemia 2010; 24: 1065–1069.
Reindl C, Quentmeier H, Petropoulos K, Greif PA, Benthaus T, Argiropoulos B et al. CBL exon 8/9 mutants activate the FLT3 pathway and cluster in core binding factor/11q deletion acute myeloid leukemia/myelodysplastic syndrome subtypes. Clin Cancer Res 2009; 15: 2238–2247.
Abbas S, Rotmans G, Lowenberg B, Valk PJ . Exon 8 splice site mutations in the gene encoding the E3-ligase CBL are associated with core binding factor acute myeloid leukemias. Haematologica 2008; 93: 1595–1597.
Becker H, Yoshida K, Blagitko-Dorfs N, Claus R, Pantic M, Abdelkarim M et al. Tracing the development of acute myeloid leukemia in CBL syndrome. Blood 2014; 123: 1883–1886.
Dunbar AJ, Gondek LP, O'Keefe CL, Makishima H, Rataul MS, Szpurka H et al. 250K single nucleotide polymorphism array karyotyping identifies acquired uniparental disomy and homozygous mutations, including novel missense substitutions of c-Cbl, in myeloid malignancies. Cancer Res 2008; 68: 10349–10357.
Grand FH, Hidalgo-Curtis CE, Ernst T, Zoi K, Zoi C, McGuire C et al. Frequent CBL mutations associated with 11q acquired uniparental disomy in myeloproliferative neoplasms. Blood 2009; 113: 6182–6192.
Sanada M, Suzuki T, Shih LY, Otsu M, Kato M, Yamazaki S et al. Gain-of-function of mutated C-CBL tumour suppressor in myeloid neoplasms. Nature 2009; 460: 904–908.
Muramatsu H, Makishima H, Jankowska AM, Cazzolli H, O'Keefe C, Yoshida N et al. Mutations of an E3 ubiquitin ligase c-Cbl but not TET2 mutations are pathogenic in juvenile myelomonocytic leukemia. Blood 2010; 115: 1969–1975.
Loh ML, Sakai DS, Flotho C, Kang M, Fliegauf M, Archambeault S et al. Mutations in CBL occur frequently in juvenile myelomonocytic leukemia. Blood 2009; 114: 1859–1863.
Makishima H, Cazzolli H, Szpurka H, Dunbar A, Tiu R, Huh J et al. Mutations of e3 ubiquitin ligase cbl family members constitute a novel common pathogenic lesion in myeloid malignancies. J Clin Oncol 2009; 27: 6109–6116.
Kohlmann A, Grossmann V, Klein HU, Schindela S, Weiss T, Kazak B et al. Next-generation sequencing technology reveals a characteristic pattern of molecular mutations in 72.8% of chronic myelomonocytic leukemia by detecting frequent alterations in TET2, CBL, RAS, and RUNX1. J Clin Oncol 2010; 28: 3858–3865.
Schnittger S, Bacher U, Alpermann T, Reiter A, Ulke M, Dicker F et al. Use of CBL exon 8 and 9 mutations in diagnosis of myeloproliferative neoplasms and myeloproliferative/myelodysplastic disorders: an analysis of 636 cases. Haematologica 2012; 97: 1890–1894.
Niemeyer CM, Kang MW, Shin DH, Furlan I, Erlacher M, Bunin NJ et al. Germline CBL mutations cause developmental abnormalities and predispose to juvenile myelomonocytic leukemia. Nat Genet 2010; 42: 794–800.
Sargin B, Choudhary C, Crosetto N, Schmidt MH, Grundler R, Rensinghoff M et al. Flt3-dependent transformation by inactivating c-Cbl mutations in AML. Blood 2007; 110: 1004–1012.
Levkowitz G, Waterman H, Ettenberg SA, Katz M, Tsygankov AY, Alroy I et al. Ubiquitin ligase activity and tyrosine phosphorylation underlie suppression of growth factor signaling by c-Cbl/Sli-1. Mol Cell 1999; 4: 1029–1040.
Saur SJ, Sangkhae V, Geddis AE, Kaushansky K, Hitchcock IS . Ubiquitination and degradation of the thrombopoietin receptor c-Mpl. Blood 2010; 115: 1254–1263.
Yokouchi M, Kondo T, Sanjay A, Houghton A, Yoshimura A, Komiya S et al. Src-catalyzed phosphorylation of c-Cbl leads to the interdependent ubiquitination of both proteins. J Biol Chem 2001; 276: 35185–35193.
Zeng S, Xu Z, Lipkowitz S, Longley JB . Regulation of stem cell factor receptor signaling by Cbl family proteins (Cbl-b/c-Cbl). Blood 2005; 105: 226–232.
Ogawa S, Shih LY, Suzuki T, Otsu M, Nakauchi H, Koeffler HP et al. Deregulated intracellular signaling by mutated c-CBL in myeloid neoplasms. Clin Cancer Res 2010; 16: 3825–3831.
Schmidt MH, Dikic I . The Cbl interactome and its functions. Nat Rev Mol Cell Biol 2005; 6: 907–918.
Ryan PE, Davies GC, Nau MM, Lipkowitz S . Regulating the regulator: negative regulation of Cbl ubiquitin ligases. Trends Biochem Sci 2006; 31: 79–88.
Kowanetz K, Crosetto N, Haglund K, Schmidt MH, Heldin CH, Dikic I . Suppressors of T-cell receptor signaling Sts-1 and Sts-2 bind to Cbl and inhibit endocytosis of receptor tyrosine kinases. J Biol Chem 2004; 279: 32786–32795.
Lee ST, Feng M, Wei Y, Li Z, Qiao Y, Guan P et al. Protein tyrosine phosphatase UBASH3B is overexpressed in triple-negative breast cancer and promotes invasion and metastasis. Proc Natl Acad Sci USA 2013; 110: 11121–11126.
Rathinam C, Thien CB, Langdon WY, Gu H, Flavell RA . The E3 ubiquitin ligase c-Cbl restricts development and functions of hematopoietic stem cells. Genes Dev 2008; 22: 992–997.
Rathinam C, Thien CB, Flavell RA, Langdon WY . Myeloid leukemia development in c-Cbl RING finger mutant mice is dependent on FLT3 signaling. Cancer Cell 2010; 18: 341–352.
Mulloy JC, Cammenga J, MacKenzie KL, Berguido FJ, Moore MA, Nimer SD . The AML1-ETO fusion protein promotes the expansion of human hematopoietic stem cells. Blood 2002; 99: 15–23.
Mulloy JC, Cammenga J, Berguido FJ, Wu K, Zhou P, Comenzo RL et al. Maintaining the self-renewal and differentiation potential of human CD34+ hematopoietic cells using a single genetic element. Blood 2003; 102: 4369–4376.
Wunderlich M, Krejci O, Wei J, Mulloy JC . Human CD34+ cells expressing the inv(16) fusion protein exhibit a myelomonocytic phenotype with greatly enhanced proliferative ability. Blood 2006; 108: 1690–1697.
Wunderlich M, Chou FS, Link KA, Mizukawa B, Perry RL, Carroll M et al. AML xenograft efficiency is significantly improved in NOD/SCID-IL2RG mice constitutively expressing human SCF, GM-CSF and IL-3. Leukemia 2010; 24: 1785–1788.
Wunderlich M, Mulloy JC . Model systems for examining effects of leukemia-associated oncogenes in primary human CD34+ cells via retroviral transduction. Methods Mol Biol 2009; 538: 263–285.
Mulloy JC, Wunderlich M, Zheng Y, Wei J . Transforming human blood stem and progenitor cells: a new way forward in leukemia modeling. Cell Cycle 2008; 7: 3314–3319.
Chen P, Price C, Li Z, Li Y, Cao D, Wiley A et al. miR-9 is an essential oncogenic microRNA specifically overexpressed in mixed lineage leukemia-rearranged leukemia. Proc Natl Acad Sci USA 2013; 110: 11511–11516.
Chou FS, Griesinger A, Wunderlich M, Lin S, Link KA, Shrestha M et al. The THPO/MPL/Bcl-xL pathway is essential for survival and self-renewal in human pre-leukemia induced by AML1-ETO. Blood 2012; 120: 709–719.
Krejci O, Wunderlich M, Geiger H, Chou FS, Schleimer D, Jansen M et al. p53 signaling in response to increased DNA damage sensitizes AML1-ETO cells to stress-induced death. Blood 2008; 111: 2190–2199.
Ptasinska A, Assi SA, Mannari D, James SR, Williamson D, Dunne J et al. Depletion of RUNX1/ETO in t(8;21) AML cells leads to genome-wide changes in chromatin structure and transcription factor binding. Leukemia 2012; 26: 1829–1841.
Ptasinska A, Assi SA, Martinez-Soria N, Imperato MR, Piper J, Cauchy P et al. Identification of a dynamic core transcriptional network in t(8;21) AML that regulates differentiation block and self-renewal. Cell Rep 2014; 8: 1974–1988.
Bagger FO, Rapin N, Theilgaard-Monch K, Kaczkowski B, Thoren LA, Jendholm J et al. HemaExplorer: a database of mRNA expression profiles in normal and malignant haematopoiesis. Nucleic Acids Res 2013; 41: D1034–D1039.
Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013; 6: pl1.
Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012; 2: 401–404.
Yang JH, Li JH, Shao P, Zhou H, Chen YQ, Qu LH . starBase: a database for exploring microRNA-mRNA interaction maps from Argonaute CLIP-Seq and Degradome-Seq data. Nucleic Acids Res 2011; 39: D202–D209.
Li JH, Liu S, Zhou H, Qu LH, Yang JH . starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res 2014; 42: D92–D97.
Lewis BP, Burge CB, Bartel DP . Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005; 120: 15–20.
Goyama S, Schibler J, Cunningham L, Zhang Y, Rao Y, Nishimoto N et al. Transcription factor RUNX1 promotes survival of acute myeloid leukemia cells. J Clin Invest 2013; 123: 3876–3888.
Wei J, Wunderlich M, Fox C, Alvarez S, Cigudosa JC, Wilhelm JS et al. Microenvironment determines lineage fate in a human model of MLL-AF9 leukemia. Cancer Cell 2008; 13: 483–495.
Asou H, Tashiro S, Hamamoto K, Otsuji A, Kita K, Kamada N . Establishment of a human acute myeloid leukemia cell line (Kasumi-1) with 8;21 chromosome translocation. Blood 1991; 77: 2031–2036.
Larizza L, Magnani I, Beghini A . The Kasumi-1 cell line: a t(8;21)-kit mutant model for acute myeloid leukemia. Leuk Lymphoma 2005; 46: 247–255.
Chou FS, Griesinger A, Wunderlich M, Lin S, Link KA, Shrestha M et al. The thrombopoietin/MPL/Bcl-xL pathway is essential for survival and self-renewal in human preleukemia induced by AML1-ETO. Blood 2012; 120: 709–719.
Sanjay A, Houghton A, Neff L, DiDomenico E, Bardelay C, Antoine E et al. Cbl associates with Pyk2 and Src to regulate Src kinase activity, alpha(v)beta(3) integrin-mediated signaling, cell adhesion, and osteoclast motility. J Cell Biol 2001; 152: 181–195.
Zhang H, Alberich-Jorda M, Amabile G, Yang H, Staber PB, Di Ruscio A et al. Sox4 is a key oncogenic target in C/EBPalpha mutant acute myeloid leukemia. Cancer Cell 2013; 24: 575–588.
Mendler JH, Maharry K, Radmacher MD, Mrozek K, Becker H, Metzeler KH et al. RUNX1 mutations are associated with poor outcome in younger and older patients with cytogenetically normal acute myeloid leukemia and with distinct gene and MicroRNA expression signatures. J Clin Oncol 2012; 30: 3109–3118.
Emmrich S, Katsman-Kuipers JE, Henke K, Khatib ME, Jammal R, Engeland F et al. miR-9 is a tumor suppressor in pediatric AML with t(8;21). Leukemia 2014; 28: 1022–1032.
Mikhailik A, Ford B, Keller J, Chen Y, Nassar N, Carpino N . A phosphatase activity of Sts-1 contributes to the suppression of TCR signaling. Mol Cell 2007; 27: 486–497.
Thomas DH, Getz TM, Newman TN, Dangelmaier CA, Carpino N, Kunapuli SP et al. A novel histidine tyrosine phosphatase, TULA-2, associates with Syk and negatively regulates GPVI signaling in platelets. Blood 2010; 116: 2570–2578.
Lo MC, Peterson LF, Yan M, Cong X, Hickman JH, Dekelver RC et al. JAK inhibitors suppress t(8;21) fusion protein-induced leukemia. Leukemia 2013; 27: 2272–2279.
Krauth MT, Eder C, Alpermann T, Bacher U, Nadarajah N, Kern W et al. High number of additional genetic lesions in acute myeloid leukemia with t(8;21)/RUNX1-RUNX1T1: frequency and impact on clinical outcome. Leukemia 2014; 28: 1449–1458.
Wang YY, Zhou GB, Yin T, Chen B, Shi JY, Liang WX et al. AML1-ETO and C-KIT mutation/overexpression in t(8;21) leukemia: implication in stepwise leukemogenesis and response to Gleevec. Proc Natl Acad Sci USA 2005; 102: 1104–1109.
Wang YY, Zhao LJ, Wu CF, Liu P, Shi L, Liang Y et al. C-KIT mutation cooperates with full-length AML1-ETO to induce acute myeloid leukemia in mice. Proc Natl Acad Sci USA 2011; 108: 2450–2455.
Wichmann C, Quagliano-Lo Coco I, Yildiz O, Chen-Wichmann L, Weber H, Syzonenko T et al. Activating c-KIT mutations confer oncogenic cooperativity and rescue RUNX1/ETO-induced DNA damage and apoptosis in human primary CD34+ hematopoietic progenitors. Leukemia 2014; 29: 279–289, 4.
Pulikkan JA, Madera D, Xue L, Bradley P, Landrette SF, Kuo YH et al. Thrombopoietin/MPL participates in initiating and maintaining RUNX1-ETO acute myeloid leukemia via PI3K/AKT signaling. Blood 2012; 120: 868–879.
Nick HJ, Kim HG, Chang CW, Harris KW, Reddy V, Klug CA . Distinct classes of c-Kit-activating mutations differ in their ability to promote RUNX1-ETO-associated acute myeloid leukemia. Blood 2012; 119: 1522–1531.
Taylor SJ, Dagger SA, Thien CB, Wikstrom ME, Langdon WY . Flt3 inhibitor AC220 is a potent therapy in a mouse model of myeloproliferative disease driven by enhanced wild-type Flt3 signaling. Blood 2012; 120: 4049–4057.
Makishima H, Sugimoto Y, Szpurka H, Clemente MJ, Ng KP, Muramatsu H et al. CBL mutation-related patterns of phosphorylation and sensitivity to tyrosine kinase inhibitors. Leukemia 2012; 26: 1547–1554.
Bunda S, Kang MW, Sybingco SS, Weng J, Favre H, Shin DH et al. Inhibition of SRC corrects GM-CSF hypersensitivity that underlies juvenile myelomonocytic leukemia. Cancer Res 2013; 73: 2540–2550.
Gilliland DG, Jordan CT, Felix CA . The molecular basis of leukemia. Hematology Am Soc Hematol Educ Program 2004, 80–97.
Schessl C, Rawat VP, Cusan M, Deshpande A, Kohl TM, Rosten PM et al. The AML1-ETO fusion gene and the FLT3 length mutation collaborate in inducing acute leukemia in mice. J Clin Invest 2005; 115: 2159–2168.
Zhao L, Melenhorst JJ, Alemu L, Kirby M, Anderson S, Kench M et al. KIT with D816 mutations cooperates with CBFB-MYH11 for leukemogenesis in mice. Blood 2012; 119: 1511–1521.
Chou FS, Wunderlich M, Griesinger A, Mulloy JC . N-Ras(G12D) induces features of stepwise transformation in preleukemic human umbilical cord blood cultures expressing the AML1-ETO fusion gene. Blood 2011; 117: 2237–2240.
Micol JB, Duployez N, Boissel N, Petit A, Geffroy S, Nibourel O et al. Frequent ASXL2 mutations in acute myeloid leukemia patients with t(8;21)/RUNX1-RUNX1T1 chromosomal translocations. Blood 2014; 124: 1445–1449.
Ellegast JM, Saito Y, Flavell RA, Manz MG . MISTRG mice support good-risk AML engraftment. ASH 2014 Annual Meeting Abstracts 2014, p. 3808.
Rongvaux A, Willinger T, Martinek J, Strowig T, Gearty SV, Teichmann LL et al. Development and function of human innate immune cells in a humanized mouse model. Nat Biotechnol 2014; 32: 364–372.
Rongvaux A, Willinger T, Takizawa H, Rathinam C, Auerbach W, Murphy AJ et al. Human thrombopoietin knockin mice efficiently support human hematopoiesis in vivo. Proc Natl Acad Sci USA 2011; 108: 2378–2383.
Acknowledgements
We thank Dr Seishi Ogawa, Dr Masashi Sanada, Dr Masafumi Onodera and Dr Karsten Spiekermann for plasmids. We thank the Flow Cytometry Core and the Mouse Core at Cincinnati Children's Hospital Medical Center for their help. This work was supported by a grant from the CancerFree Kids Foundation for Cancer Research (JCM and SG), an Institutional Clinical and Translational Science Award, NIH/NCRR Grant Number 1UL1RR026314-01, Translational Trials Development and Support Laboratory award (U.S.P.H.S. Grant Number MO1 RR 08084), a Center of Excellence in Molecular Hematology P30 award (DK090971), JSPS Postdoctoral Fellowship for Research Abroad (SG), CA178454 (JC), the Coleman Leukemia Research Foundation (CDB), CA180861 (CDB), CA101140 (CDB), CA140158 (CDB), grants from Leukaemia Lymphoma Research (12007, CB) and (12055, OH). AG and NNN were supported by a grant from the Leukemia and Lymphoma Society. JCM is a Leukemia and Lymphoma Society Scholar.
Author contributions
SG conceived the project, designed and performed the research, analyzed the data and wrote the paper. JS performed the research and analyzed the data. MS, SL and KAL assisted with experiments. AG and NNN actively participated in designing and assisting with the experiments regarding UBASH3B/Sts-1. JC assisted with the experiments regarding miR-9. SPW, CDB and DN analyzed expression profiles of CBF-AML patients. SA, AP, OH and CB performed ChIP-Seq and RNA-Seq analyses. TK analyzed the data. JCM secured funding, analyzed the data and participated in writing the paper.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Leukemia website
Supplementary information
Rights and permissions
About this article
Cite this article
Goyama, S., Schibler, J., Gasilina, A. et al. UBASH3B/Sts-1-CBL axis regulates myeloid proliferation in human preleukemia induced by AML1-ETO. Leukemia 30, 728–739 (2016). https://doi.org/10.1038/leu.2015.275
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2015.275
This article is cited by
-
Hyperactive Natural Killer cells in Rag2 knockout mice inhibit the development of acute myeloid leukemia
Communications Biology (2023)
-
Non-proteolytic ubiquitylation in cellular signaling and human disease
Communications Biology (2022)
-
Antitumor immunity augments the therapeutic effects of p53 activation on acute myeloid leukemia
Nature Communications (2019)
-
Opposing effects of acute versus chronic inhibition of p53 on decitabine’s efficacy in myeloid neoplasms
Scientific Reports (2019)
-
Subtype-specific regulatory network rewiring in acute myeloid leukemia
Nature Genetics (2019)