Abstract
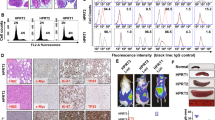
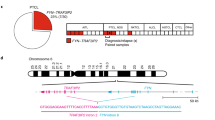
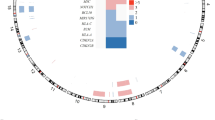
Although anaplastic large-cell lymphomas (ALCL) carrying anaplastic lymphoma kinase (ALK) have a relatively good prognosis, aggressive forms exist. We have identified a novel translocation, causing the fusion of the TRAF1 and ALK genes, in one patient who presented with a leukemic ALK+ ALCL (ALCL-11). To uncover the mechanisms leading to high-grade ALCL, we developed a human patient-derived tumorgraft (hPDT) line. Molecular characterization of primary and PDT cells demonstrated the activation of ALK and nuclear factor kB (NFkB) pathways. Genomic studies of ALCL-11 showed the TP53 loss and the in vivo subclonal expansion of lymphoma cells, lacking PRDM1/Blimp1 and carrying c-MYC gene amplification. The treatment with proteasome inhibitors of TRAF1-ALK cells led to the downregulation of p50/p52 and lymphoma growth inhibition. Moreover, a NFkB gene set classifier stratified ALCL in distinct subsets with different clinical outcome. Although a selective ALK inhibitor (CEP28122) resulted in a significant clinical response of hPDT mice, nevertheless the disease could not be eradicated. These data indicate that the activation of NFkB signaling contributes to the neoplastic phenotype of TRAF1-ALK ALCL. ALCL hPDTs are invaluable tools to validate the role of druggable molecules, predict therapeutic responses and implement patient specific therapies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Falini B, Martelli MP . Anaplastic large cell lymphoma: changes in the World Health Organization classification and perspectives for targeted therapy. Haematologica 2009; 94: 897–900.
Swerdlow SH CE, Haris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Vardiman JW . WHO classification of tumors of haemotolopoietic and lymphoid tissues, 4th edn. Stylus Publishing, LLC: Sterling VA, 2008.
Savage KJ, Harris NL, Vose JM, Ullrich F, Jaffe ES, Connors JM et al. ALK- anaplastic large-cell lymphoma is clinically and immunophenotypically different from both ALK+ ALCL and peripheral T-cell lymphoma, not otherwise specified: report from the International Peripheral T-Cell Lymphoma Project. Blood 2008; 111: 5496–5504.
Schmitz N, Trumper L, Ziepert M, Nickelsen M, Ho AD, Metzner B et al. Treatment and prognosis of mature T-cell and NK-cell lymphoma: an analysis of patients with T-cell lymphoma treated in studies of the German High-Grade Non-Hodgkin Lymphoma Study Group. Blood 2010; 116: 3418–3425.
Monaco S, Tsao L, Murty VV, Nandula SV, Donovan V, Oesterheld J et al. Pediatric ALK+ anaplastic large cell lymphoma with t(3;8)(q26.2;q24) translocation and c-myc rearrangement terminating in a leukemic phase. Am J Hematol 2007; 82: 59–64.
Liang X, Branchford B, Greffe B, McGavran L, Carstens B, Meltesen L et al. Dual ALK and MYC Rearrangements Leading to an Aggressive Variant of Anaplastic Large Cell Lymphoma. J Pediatr Oncol 2013; 35: e209–e213.
Ferreri AJ, Govi S, Pileri SA, Savage KJ . Anaplastic large cell lymphoma, ALK-positive. Crit Rev Oncology Hematol 2012; 83: 293–302.
Barreca A, Lasorsa E, Riera L, Machiorlatti R, Piva R, Ponzoni M et al. Anaplastic lymphoma kinase in human cancer. J Mol Endocrinol 2011; 47: R11–R23.
Miranda C, Roccato E, Raho G, Pagliardini S, Pierotti MA, Greco A . The TFG protein, involved in oncogenic rearrangements, interacts with TANK and NEMO, two proteins involved in the NF-kappaB pathway. J Cell Physiol 2006; 208: 154–160.
Boi M, Rinaldi A, Kwee I, Bonetti P, Todaro M, Tabbo F et al. PRDM1/BLIMP1 is commonly inactivated in anaplastic large T-cell lymphoma. Blood 2013; 122: 2683–2693.
Grewal JS, Smith LB, Winegarden JD 3rd, Krauss JC, Tworek JA, Schnitzer B . Highly aggressive ALK-positive anaplastic large cell lymphoma with a leukemic phase and multi-organ involvement: a report of three cases and a review of the literature. Ann Hematol 2007; 86: 499–508.
Moritake H, Shimonodan H, Marutsuka K, Kamimura S, Kojima H, Nunoi H . C-MYC rearrangement may induce an aggressive phenotype in anaplastic lymphoma kinase positive anaplastic large cell lymphoma: Identification of a novel fusion gene ALO17/C-MYC. Am J Hematol 2011; 86: 75–78.
Shultz LD, Brehm MA, Garcia-Martinez JV, Greiner DL . Humanized mice for immune system investigation: progress, promise and challenges. Nat Rev Immunol 2012; 12: 786–798.
Quintana E, Shackleton M, Sabel MS, Fullen DR, Johnson TM, Morrison SJ . Efficient tumour formation by single human melanoma cells. Nature 2008; 456: 593–598.
Tentler JJ, Tan AC, Weekes CD, Jimeno A, Leong S, Pitts TM et al. Patient-derived tumour xenografts as models for oncology drug development. Nat Rev Clin Oncol 2012; 9: 338–350.
Garber K . From human to mouse and back: 'tumorgraft' models surge in popularity. J Natl Cancer Inst 2009; 101: 6–8.
Vilas-Zornoza A, Agirre X, Abizanda G, Moreno C, Segura V, De Martino Rodriguez A et al. Preclinical activity of LBH589 alone or in combination with chemotherapy in a xenogeneic mouse model of human acute lymphoblastic leukemia. Leukemia 2012; 26: 1517–1526.
Cheng M, Quail MR, Gingrich DE, Ott GR, Lu L, Wan W et al. CEP-28122, a highly potent and selective orally active inhibitor of anaplastic lymphoma kinase with antitumor activity in experimental models of human cancers. Mol Cancer Ther 2011; 11: 670–679.
Rubio-Viqueira B, Jimeno A, Cusatis G, Zhang X, Iacobuzio-Donahue C, Karikari C et al. An in vivo platform for translational drug development in pancreatic cancer. Clinical Cancer Res 2006; 12: 4652–4661.
Feldman AL, Vasmatzis G, Asmann YW, Davila J, Middha S, Eckloff BW et al. Novel TRAF1-ALK fusion identified by deep RNA sequencing of anaplastic large cell lymphoma. Genes Chromosomes Cancer 2013; 52: 1097–1102.
Tabbo F, Barreca A, Machiorlatti R, Messana K, Landra I, Abate F et al. Humanized NOD/Scid/IL2g−/− tumor grafts recapitulate primary anaplastic large cell lymphoma. AACR Annual Meeting 2013 2013; (abstract 3853).
Piva R, Agnelli L, Pellegrino E, Todoerti K, Grosso V, Tamagno I et al. Gene expression profiling uncovers molecular classifiers for the recognition of anaplastic large-cell lymphoma within peripheral T-cell neoplasms. J Clin Oncol 2010; 28: 1583–1590.
Palomero T, Couronne L, Khiabanian H, Kim MY, Ambesi-Impiombato A, Perez-Garcia A et al. Recurrent mutations in epigenetic regulators, RHOA and FYN kinase in peripheral T cell lymphomas. Nat Genet 2014; 46: 166–170.
Abate F, Acquaviva A, Paciello G, Foti C, Ficarra E, Ferrarini A et al. Bellerophontes: an RNA-Seq data analysis framework for chimeric transcripts discovery based on accurate fusion model. Bioinformatics 2012; 28: 2114–2121.
McPherson A, Hormozdiari F, Zayed A, Giuliany R, Ha G, Sun MG et al. deFuse: an algorithm for gene fusion discovery in tumor RNA-Seq data. PLoS Comput Biol 2011; 7: e1001138.
Iyer MK, Chinnaiyan AM, Maher C . ChimeraScan: a tool for identifying chimeric transcription in sequencing data. Bioinformatics 2011; 27: 2903–2904.
Singh D, Chan JM, Zoppoli P, Niola F, Sullivan R, Castano A et al. Transforming fusions of FGFR and TACC genes in human glioblastoma. Science 2012; 337: 1231–1235.
Ma Z, Cools J, Marynen P, Cui X, Siebert R, Gesk S et al. Inv(2)(p23q35) in anaplastic large-cell lymphoma induces constitutive anaplastic lymphoma kinase (ALK) tyrosine kinase activation by fusion to ATIC, an enzyme involved in purine nucleotide biosynthesis. Blood 2000; 95: 2144–2149.
Agnelli L, Mereu E, Pellegrino E, Limongi T, Kwee I, Bergaggio E et al. Identification of a 3-gene model as a powerful diagnostic tool for the recognition of ALK-negative anaplastic large-cell lymphoma. Blood 2012; 120: 1274–1281.
Shultz LD, Ishikawa F, Greiner DL . Humanized mice in translational biomedical research. Nat Rev Immunology 2007; 7: 118–130.
Brusa D, Serra S, Coscia M, Rossi D, D'Arena G, Laurenti L et al. The PD-1/PD-L1 axis contributes to T-cell dysfunction in chronic lymphocytic leukemia. Haematologica 2013; 98: 953–963.
Zamo A, Chiarle R, Piva R, Howes J, Fan Y, Chilosi M et al. Anaplastic lymphoma kinase (ALK) activates Stat3 and protects hematopoietic cells from cell death. Oncogene 2002; 21: 1038–1047.
Piva R, Chiarle R, Manazza AD, Taulli R, Simmons W, Ambrogio C et al. Ablation of oncogenic ALK is a viable therapeutic approach for anaplastic large-cell lymphomas. Blood 2006; 107: 689–697.
Boccalatte FE, Voena C, Riganti C, Bosia A, D'Amico L, Riera L et al. The enzymatic activity of 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase is enhanced by NPM-ALK: new insights in ALK-mediated pathogenesis and the treatment of ALCL. Blood 2009; 113: 2776–2790.
Fu L, Medico E . FLAME, a novel fuzzy clustering method for the analysis of DNA microarray data. BMC Bioinformatics 2007; 8: 3.
Iqbal J, Wright G, Wang C, Rosenwald A, Gascoyne RD, Weisenburger DD et al. Gene expression signatures delineate biological and prognostic subgroups in peripheral T-cell lymphoma. Blood 2014; 123: 2915–2923.
Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP . GenePattern 2.0. Nat Genet 2006; 38: 500–501.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Nat l Acad Sci USA 2005; 102: 15545–15550.
Jiang C, Xuan Z, Zhao F, Zhang MQ . TRED: a transcriptional regulatory element database, new entries and other development. Nucleic Acids Res 2007; 35: D137–D140.
Feuerhake F, Kutok JL, Monti S, Chen W, LaCasce AS, Cattoretti G et al. NFkappaB activity, function, and target-gene signatures in primary mediastinal large B-cell lymphoma and diffuse large B-cell lymphoma subtypes. Blood 2005; 106: 1392–1399.
Lee ST, Li Z, Wu Z, Aau M, Guan P, Karuturi RK et al. Context-specific regulation of NF-kappaB target gene expression by EZH2 in breast cancers. Mol Cell 2011; 43: 798–810.
Nagar M, Jacob-Hirsch J, Vernitsky H, Berkun Y, Ben-Horin S, Amariglio N et al. TNF activates a NF-kappaB-regulated cellular program in human CD45RA- regulatory T cells that modulates their suppressive function. J Immunol 2010; 184: 3570–3581.
Rinaldi A, Kwee I, Young KH, Zucca E, Gaidano G, Forconi F et al. Genome-wide high resolution DNA profiling of hairy cell leukaemia. Br J Haematol 2013; 162: 566–569.
Cools J, Wlodarska I, Somers R, Mentens N, Pedeutour F, Maes B et al. Identification of novel fusion partners of ALK, the anaplastic lymphoma kinase, in anaplastic large-cell lymphoma and inflammatory myofibroblastic tumor. Genes Chromosomes Cancer 2002; 34: 354–362.
Tabbo F, Barreca A, Piva R, Inghirami G . ALK signaling and target therapy in anaplastic large cell lymphoma. Front Oncol 2012; 2: 41.
Horie R, Watanabe M, Ishida T, Koiwa T, Aizawa S, Itoh K et al. The NPM-ALK oncoprotein abrogates CD30 signaling and constitutive NF-kappaB activation in anaplastic large cell lymphoma. Cancer Cell 2004; 5: 353–364.
Hideshima T, Ikeda H, Chauhan D, Okawa Y, Raje N, Podar K et al. Bortezomib induces canonical nuclear factor-kappaB activation in multiple myeloma cells. Blood 2009; 114: 1046–1052.
Compagno M, Lim WK, Grunn A, Nandula SV, Brahmachary M, Shen Q et al. Mutations of multiple genes cause deregulation of NF-kappaB in diffuse large B-cell lymphoma. Nature 2009; 459: 717–721.
Lenz G, Wright G, Dave SS, Xiao W, Powell J, Zhao H et al. Stromal gene signatures in large-B-cell lymphomas. N Eng J Med 2008; 359: 2313–2323.
Zhang Q, Wei F, Wang HY, Liu X, Roy D, Xiong QB et al. The potent oncogene NPM-ALK mediates malignant transformation of normal human CD4(+) T lymphocytes. Am J Pathol 2013; 183: 1971–1980.
Martinez-Delgado B, Cuadros M, Honrado E, Ruiz de la Parte A, Roncador G, Alves J et al. Differential expression of NF-kappaB pathway genes among peripheral T-cell lymphomas. Leukemia 2005; 19: 2254–2263.
Bargou RC, Leng C, Krappmann D, Emmerich F, Mapara MY, Bommert K et al. High-level nuclear NF-kappa B and Oct-2 is a common feature of cultured Hodgkin/Reed-Sternberg cells. Blood 1996; 87: 4340–4347.
Mathas S, Johrens K, Joos S, Lietz A, Hummel F, Janz M et al. Elevated NF-kappaB p50 complex formation and Bcl-3 expression in classical Hodgkin, anaplastic large-cell, and other peripheral T-cell lymphomas. Blood 2005; 106: 4287–4293.
Eckerle S, Brune V, Doring C, Tiacci E, Bohle V, Sundstrom C et al. Gene expression profiling of isolated tumour cells from anaplastic large cell lymphomas: insights into its cellular origin, pathogenesis and relation to Hodgkin lymphoma. Leukemia 2009; 23: 2129–2138.
Hiruma Y, Honjo T, Jelinek DF, Windle JJ, Shin J, Roodman GD et al. Increased signaling through p62 in the marrow microenvironment increases myeloma cell growth and osteoclast formation. Blood 2009; 113: 4894–4902.
Lwin T, Hazlehurst LA, Li Z, Dessureault S, Sotomayor E, Moscinski LC et al. Bone marrow stromal cells prevent apoptosis of lymphoma cells by upregulation of anti-apoptotic proteins associated with activation of NF-kappaB (RelB/p52) in non-Hodgkin's lymphoma cells. Leukemia 2007; 21: 1521–1531.
Laimer D, Dolznig H, Kollmann K, Vesely PW, Schlederer M, Merkel O et al. PDGFR blockade is a rational and effective therapy for NPM-ALK-driven lymphomas. Nat Med 2012; 18: 1699–1704.
Vasmatzis G, Johnson SH, Knudson RA, Ketterling RP, Braggio E, Fonseca R et al. Genome-wide analysis reveals recurrent structural abnormalities of TP63 and other p53-related genes in peripheral T-cell lymphomas. Blood 2012; 120: 2280–2289.
Clappier E, Gerby B, Sigaux F, Delord M, Touzri F, Hernandez L et al. Clonal selection in xenografted human T cell acute lymphoblastic leukemia recapitulates gain of malignancy at relapse. J Exp Med 2011; 208: 653–661.
Acknowledgements
GI is supported by the Italian Association for Cancer Research Special Program in Clinical Molecular Oncology, Milan (5 × 1000 No. 10007); Regione Piemonte (ONCOPROT, CIPE 25/2005); ImmOnc (Innovative approaches to boost the immune responses, Programma Operativo Regionale, Piattaforme Innovative BIO FESR 2007/13, Asse 1 ‘Ricerca e innovazione’ della LR 34/2004) and the Oncology Program of Compagnia di San Paolo, Torino. SP and BF are supported by the Italian Association for Cancer Research Special Program in Clinical Molecular Oncology, Milan (5x1000 No. 10007); RR by Partnership for Cure, NIH 1 P50 MH094267-01, NIH 1 U54 CA121852-05, NIH 1R01CA164152-01. FB is sponsored by the Oncosuisse KLS-02403-02-2009 (Bern, Switzerland); Anna Lisa Stiftung (Ascona, Switzerland); Nelia and Amadeo Barletta Foundation (Lausanne, Switzerland); RP by Rete Oncologica del Piemonte e della Valle d’Aosta. LDS is sponsored by National Institutes of Health (USA) grant CA034196. MB, MT, IL, FT FDG, RC and LB are enrolled in the PhD program (Pharmaceutical Sciences, University of Geneva, Switzerland and Molecular Medicine, University of Torino, respectively). We thank Drs Vigliani C, Fioravanti A and Mossino M for their technical support and Dr Casano J for the constructive revision of the manuscript.
Author information
Authors and Affiliations
Consortia
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Leukemia website
Supplementary information
Appendix
Appendix
Appendix The European T-Cell Lymphoma Study Group:
Italy: Cristina Abele, Luca Bessone, Antonella Barreca, Michela Boi, Federica Cavallo, Nicoletta Chiesa, Ramona Crescenzo, Antonella Fienga, Marcello Gaudiano, Filomena di Giacomo, Giorgio Inghirami, Indira Landra, Elena Lasorsa, Rodolfo Marchiorlatti, Barbara Martinoglio, Enzo Medico, Gian Battista Ferrero, Katia Messana, Elisabetta Mereu, Elisa Pellegrino, Roberto Piva, Irene Scafò, Elisa Spaccarotella, Fabrizio Tabbò, Maria Todaro, Ivana Ubezzi, Susanna Urigu (Azienda Ospedaliera Città della Salute e della Scienza di Torino, University of and Center for Experimental Research and Medical Studies); Domenico Novero, Annalisa Chiapella and Umberto Vitolo (Azienda Ospedaliera Città della Salute e della Scienza di Torino and San Luigi Gonzaga, Turin); Francesco Abate, Elisa Ficarra, Andrea Acquaviva (Politecnico di Torino); Luca Agnelli and Antonino Neri (University of Milan); Anna Caliò Marco Chilosi and Alberto Zamó (University of Verona); Fabio Facchetti and Silvia Lonardi (University of Brescia); Anna De Chiara and Franco Fulciniti (National Cancer Institute, Naples); Andrés Ferreri and Maurilio Ponzoni (San Raffaele Institute, Milan); Claudio Agostinelli, Pier Paolo Piccaluga and Stefano Pileri (University of Bologna); Brunangelo Falini and Enrico Tiacci (University of Perugia). Belgium: Peter Van Loo, Thomas Tousseyn, and Christiane De Wolf-Peeters (University of Leuven); Germany: Eva Geissinger, Hans Konrad Muller-Hermelink and Andreas Rosenwald, (University of Wuerzburg); Spain: Miguel Angel Piris and Maria E. Rodriguez (Hospital Universitario Marqués de Valdecilla, IFIMAV, Santander and Instituto de Investigaciones Biomédicas Alberto Sols, CSIC-UAM, Madrid); Switzerland: Francesco Bertoni, Andrea Rinaldi, Ivo Kwee (Institute of Oncology Research, Bellizona). Brasil: Carlos Chiattone (Disciplina de Hematologia e Oncologia, FCM da Santa Casa de São Paulo) and Roberto Antonio Pinto Paes (Department of Pathology FCM da Santa Casa de São Paulo).
Rights and permissions
About this article
Cite this article
Abate, F., Todaro, M., van der Krogt, JA. et al. A novel patient-derived tumorgraft model with TRAF1-ALK anaplastic large-cell lymphoma translocation. Leukemia 29, 1390–1401 (2015). https://doi.org/10.1038/leu.2014.347
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2014.347
This article is cited by
-
RNA demethylase ALKBH5 promotes tumorigenesis in multiple myeloma via TRAF1-mediated activation of NF-κB and MAPK signaling pathways
Oncogene (2022)
-
A novel model of alternative NF-κB pathway activation in anaplastic large cell lymphoma
Leukemia (2021)
-
Molecular insights into pathogenesis and targeted therapy of peripheral T cell lymphoma
Experimental Hematology & Oncology (2020)
-
Crystal structure of TRAF1 TRAF domain and its implications in the TRAF1-mediated intracellular signaling pathway
Scientific Reports (2016)