Abstract
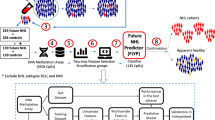
Prospective identification of patients with chronic lymphocytic leukemia (CLL) destined to progress would greatly facilitate their clinical management. Recently, whole-genome DNA methylation analyses identified three clinicobiologic CLL subgroups with an epigenetic signature related to different normal B-cell counterparts. Here, we developed a clinically applicable method to identify these subgroups and to study their clinical relevance. Using a support vector machine approach, we built a prediction model using five epigenetic biomarkers that was able to classify CLL patients accurately into the three subgroups, namely naive B-cell-like, intermediate and memory B-cell-like CLL. DNA methylation was quantified by highly reproducible bisulfite pyrosequencing assays in two independent CLL series. In the initial series (n=211), the three subgroups showed differential levels of IGHV (immunoglobulin heavy-chain locus) mutation (P<0.001) and VH usage (P<0.03), as well as different clinical features and outcome in terms of time to first treatment (TTT) and overall survival (P<0.001). A multivariate Cox model showed that epigenetic classification was the strongest predictor of TTT (P<0.001) along with Binet stage (P<0.001). These findings were corroborated in a validation series (n=97). In this study, we developed a simple and robust method using epigenetic biomarkers to categorize CLLs into three subgroups with different clinicobiologic features and outcome.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Rozman C, Montserrat E . Chronic lymphocytic leukemia. NEngl J Med 1995; 333: 1052–1057.
Damle RN, Wasil T, Fais F, Ghiotto F, Valetto A, Allen SL et al. Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood 1999; 94: 1840–1847.
Hamblin TJ, Davis Z, Gardiner A, Oscier DG, Stevenson FK . Unmutated Ig V(H) genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood 1999; 94: 1848–1854.
Chiorazzi N, Ferrarini M . Cellular origin(s) of chronic lymphocytic leukemia: cautionary notes and additional considerations and possibilities. Blood 2011; 117: 1781–1791.
Kulis M, Heath S, Bibikova M, Queiros AC, Navarro A, Clot G et al. Epigenomic analysis detects widespread gene-body DNA hypomethylation in chronic lymphocytic leukemia. Nat Genet 2012; 44: 1236–1242.
Seifert M, Sellmann L, Bloehdorn J, Wein F, Stilgenbauer S, Durig J et al. Cellular origin and pathophysiology of chronic lymphocytic leukemia. J Exp Med 2012; 209: 2183–2198.
Crespo M, Bosch F, Villamor N, Bellosillo B, Colomer D, Rozman M et al. ZAP-70 expression as a surrogate for immunoglobulin-variable-region mutations in chronic lymphocytic leukemia. N Engl J Med 2003; 348: 1764–1775.
Dohner H, Stilgenbauer S, Benner A, Leupolt E, Krober A, Bullinger L et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med 2000; 343: 1910–1916.
Fabbri G, Rasi S, Rossi D, Trifonov V, Khiabanian H, Ma J et al. Analysis of the chronic lymphocytic leukemia coding genome: role of NOTCH1 mutational activation. J Exp Med 2011; 208: 1389–1401.
Puente XS, Pinyol M, Quesada V, Conde L, Ordonez GR, Villamor N et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature 2011; 475: 101–105.
Quesada V, Conde L, Villamor N, Ordonez GR, Jares P, Bassaganyas L et al. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat Genet 2012; 44: 47–52.
Wang L, Lawrence MS, Wan Y, Stojanov P, Sougnez C, Stevenson K et al. SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. N Engl J Med 2011; 365: 2497–2506.
Rossi D, Rasi S, Spina V, Bruscaggin A, Monti S, Ciardullo C et al. Integrated mutational and cytogenetic analysis identifies new prognostic subgroups in chronic lymphocytic leukemia. Blood 2013; 121: 1403–1412.
Baylin SB, Jones PA . A decade of exploring the cancer epigenome—biological and translational implications. Nat Rev Cancer 2011; 11: 726–734.
Esteller M . Epigenetics in cancer. N Engl J Med 2008; 358: 1148–1159.
Heyn H, Esteller M . DNA methylation profiling in the clinic: applications and challenges. Nat Rev 2012; 13: 679–692.
Bird A . DNA methylation patterns and epigenetic memory. Genes Dev 2002; 16: 6–21.
Bird A . Perceptions of epigenetics. Nature 2007; 447: 396–398.
Du P, Zhang X, Huang CC, Jafari N, Kibbe WA, Hou L et al. Comparison of beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinform 2010; 11: 587.
Efron B, Tibshirani RJ . Improvements on cross-validation: the 632+ bootstrap method. J Am Stat Assoc 1997; 92: 548–560.
Cortes C, Vapnik V . Support-vector networks. Mach Learn 1995; 20: 273–297.
Tost J, Gut IG . DNA methylation analysis by pyrosequencing. Nat Protoc 2007; 2: 2265–2275.
Lamprecht B, Walter K, Kreher S, Kumar R, Hummel M, Lenze D et al. Derepression of an endogenous long terminal repeat activates the CSF1R proto-oncogene in human lymphoma. Nat Med 2010; 16: 571–579.
Cheson BD, Bennett JM, Rai KR, Grever MR, Kay NE, Schiffer CA et al. Guidelines for clinical protocols for chronic lymphocytic leukemia: recommendations of the National Cancer Institute-sponsored working group. Am J Hematol 1988; 29: 152–163.
Cheson BD, Bennett JM, Grever M, Kay N, Keating MJ, O'Brien S et al. National Cancer Institute-sponsored Working Group guidelines for chronic lymphocytic leukemia: revised guidelines for diagnosis and treatment. Blood 1996; 87: 4990–4997.
Hallek M, Cheson BD, Catovsky D, Caligaris-Cappio F, Dighiero G, Dohner H et al. Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: a report from the International Workshop on Chronic Lymphocytic Leukemia updating the National Cancer Institute-Working Group 1996 guidelines. Blood 2008; 111: 5446–5456.
Kaplan EL, Meier P . Non parametric estimation from incomplete observation. J Am Stat Assoc 1958; 53: 457–481.
Peto R, Pike MC . Conservatism of the approximation sigma (O-E)2-E in the logrank test for survival data or tumor incidence data. Biometrics 1973; 29: 579–584.
Cox DR . Regression models and life tables. J R Stat Soc Ser B 1972; 34: 187–220.
Klein JP, Moeschberger ML . Survival Analysis. Techniques for Censored and Truncated Data. Springer: New York, NY, USA, 1997.
Laird PW . The power and the promise of DNA methylation markers. Nat Rev Cancer 2003; 3: 253–266.
Rodriguez-Paredes M, Esteller M . Cancer epigenetics reaches mainstream oncology. Nat Med 2011; 17: 330–339.
Fernandez AF, Assenov Y, Martin-Subero JI, Balint B, Siebert R, Taniguchi H et al. A DNA methylation fingerprint of 1628 human samples. Genome Res 2012; 22: 407–419.
Gaidano G, Foa R, Dalla-Favera R . Molecular pathogenesis of chronic lymphocytic leukemia. J Clin Invest 2012; 122: 3432–3438.
Berkowska MA, Driessen GJ, Bikos V, Grosserichter-Wagener C, Stamatopoulos K, Cerutti A et al. Human memory B cells originate from three distinct germinal center-dependent and -independent maturation pathways. Blood 2011; 118: 2150–2158.
Cerutti A, Cols M, Puga I . Marginal zone B cells: virtues of innate-like antibody-producing lymphocytes. Nat Rev Immunol 2013; 13: 118–132.
Strefford JC, Sutton LA, Baliakas P, Agathangelidis A, Malcikova J, Plevova K et al. Distinct patterns of novel gene mutations in poor-prognostic stereotyped subsets of chronic lymphocytic leukemia: the case of SF3B1 and subset #2. Leukemia 2013; 27: 2196–2199.
Zenz T, Mertens D, Kuppers R, Dohner H, Stilgenbauer S . From pathogenesis to treatment of chronic lymphocytic leukaemia. Nat Rev Cancer 2010; 10: 37–50.
Abrisqueta P, Pereira A, Rozman C, Aymerich M, Gine E, Moreno C et al. Improving survival in patients with chronic lymphocytic leukemia (1980-2008): the Hospital Clinic of Barcelona experience. Blood 2009; 114: 2044–2050.
Acknowledgements
We are grateful to S Guijarro, S Martín, C Capdevila, M Sánchez, L Plà, A Heydorn and G Riesen for excellent technical assistance, and to N Villahoz and C Muro for excellent work in the coordination of the Spanish CLL Genome Consortium. We are indebted to the Hospital Clínic de Barcelona–IDIBAPS Biobank-Tumor Bank and Hematopathology Collection for the sample procurement as well as to the technical and medical staff of all the laboratories involved in the study. We are also very grateful to the patients with CLL who have participated in this study. This work was funded by the Spanish Ministry of Economy and Competitiveness (MINECO) through the Instituto de Salud Carlos III (ISCIII) and the Red Temática de Investigación del Cáncer (RTICC) of the ISCIII (RD12/0036/0036 to EC, RD12/0036/0023 to AL-G, RD12/0036/0004 to DC and RD12/0036/0067 to CL-O) and project SAF2009-08663 (JIM-S), the UK Medical Research Council (AM, SJ and MJSD) as well as the European Union’s Seventh Framework Programme through the Blueprint Consortium (grant agreement 282510 to EC and RS). CL-O is a researcher of the Botín Foundation and EC an ICREA-Academia researcher. JIM-S is supported by a Ramon y Cajal contract of the MINECO, ACQ by the Portuguese Fundação para a Ciência e a Tecnologia, MK by the Agència de Gestió d’Ajuts Universitaris i de Recerca (Generalitat de Catalunya) and JR by a Junior Excellence Research Group of the Jackstädt foundation.
Author Contributions
ACQ, MK, AN, JR, AKB, JK, CR, NR and ML-G performed experiments. ACQ, NV, AM-T, G Clot, G Castellano, SB, IS, XSP, RS, CR, AL-G and JIM-S analyzed and interpreted data. EMM-P, MP and MA performed sample preparation and quality control. NV, AM-T, SJ, AM, DC, MA, MR, JD, EG, MG-D and MJSD reviewed the pathologic and clinical data and confirmed diagnosis. CL-O, EC and JIM-S designed the study. ACQ, NV, EC, AL-G and JIM-S wrote the manuscript. All authors read and approved the final version of the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Leukemia website
Supplementary information
Rights and permissions
About this article
Cite this article
Queirós, A., Villamor, N., Clot, G. et al. A B-cell epigenetic signature defines three biologic subgroups of chronic lymphocytic leukemia with clinical impact. Leukemia 29, 598–605 (2015). https://doi.org/10.1038/leu.2014.252
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2014.252
This article is cited by
-
Time to relapse in chronic lymphocytic leukemia and DNA-methylation-based biological age
Clinical Epigenetics (2023)
-
Network analysis reveals a major role for 14q32 cluster miRNAs in determining transcriptional differences between IGHV-mutated and unmutated CLL
Leukemia (2023)
-
Molecular characterization of Richter syndrome identifies de novo diffuse large B-cell lymphomas with poor prognosis
Nature Communications (2023)
-
CALN1 hypomethylation as a biomarker for high-risk bladder cancer
BMC Urology (2022)
-
RPPA-based proteomics recognizes distinct epigenetic signatures in chronic lymphocytic leukemia with clinical consequences
Leukemia (2022)