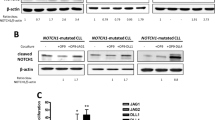
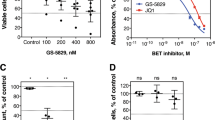
Abstract
Targeting Notch signaling has emerged as a promising therapeutic strategy for chronic lymphocytic leukemia (CLL), especially for the poor prognostic subgroup of NOTCH1-mutated patients. Here, we report that the γ-secretase inhibitor PF-03084014 inhibits the constitutive Notch activation and induces selective apoptosis in CLL cells carrying NOTCH1 mutations. Combination of PF-03084014 with fludarabine has a synergistic antileukemic effect in primary NOTCH1-mutated CLL cells, even in the presence of the protective stroma. At transcriptional level, PF-03084014 plus fludarabine treatment induces the upregulation of the proapoptotic gene HRK and the downmodulation of MMP9, IL32 and RAC2 genes that are related to invasion and chemotaxis. PF-03084014 also overcomes fludarabine-mediated activation of nuclear factor-κB signaling. Moreover, this combination impairs angiogenesis and CXCL12-induced responses in NOTCH1-mutated CLL cells, in particular those related to tumoral migration and invasion. Importantly, all these collaborative effects are specific for NOTCH1 mutation and do not occur in unmutated cases. In conclusion, we provide evidence that Notch is a therapeutic target in CLL cases with NOTCH1-activating mutations, supporting the use of Notch pathway inhibitors in combination with chemotherapy as a promising approach for the treatment of these high-risk CLL patients.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
References
Moreno C, Montserrat E . New prognostic markers in chronic lymphocytic leukemia. Blood Rev 2008; 22: 211–219.
Zenz T, Mertens D, Kuppers R, Dohner H, Stilgenbauer S . From pathogenesis to treatment of chronic lymphocytic leukaemia. Nat Rev Cancer 2010; 10: 37–50.
Puente XS, Pinyol M, Quesada V, Conde L, Ordonez GR, Villamor N et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature 2011; 475: 101–105.
Quesada V, Conde L, Villamor N, Ordonez GR, Jares P, Bassaganyas L et al. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat Genet 2012; 44: 47–52.
Ramsay AJ, Quesada V, Foronda M, Conde L, Martinez-Trillos A, Villamor N et al. POT1 mutations cause telomere dysfunction in chronic lymphocytic leukemia. Nat Genet 2013; 45: 526–530.
Wang L, Lawrence MS, Wan Y, Stojanov P, Sougnez C, Stevenson K et al. SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. N Engl J Med 2011; 365: 2497–2506.
Fabbri G, Rasi S, Rossi D, Trifonov V, Khiabanian H, Ma J et al. Analysis of the chronic lymphocytic leukemia coding genome: role of NOTCH1 mutational activation. J Exp Med 2011; 208: 1389–1401.
Rossi D, Rasi S, Fabbri G, Spina V, Fangazio M, Forconi F et al. Mutations of NOTCH1 are an independent predictor of survival in chronic lymphocytic leukemia. Blood 2012; 119: 521–529.
Weissmann S, Roller A, Jeromin S, Hernandez M, Abaigar M, Hernandez-Rivas JM et al. Prognostic impact and landscape of NOTCH1 mutations in chronic lymphocytic leukemia (CLL): a study on 852 patients. Leukemia 2013; 27: 2393–2396.
Bray SJ . Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol 2006; 7: 678–689.
Rosati E, Sabatini R, Rampino G, Tabilio A, Di IM, Fettucciari K et al. Constitutively activated Notch signaling is involved in survival and apoptosis resistance of B-CLL cells. Blood 2009; 113: 856–865.
Arruga F, Gizdic B, Serra S, Vaisitti T, Ciardullo C, Coscia M et al. Functional impact of NOTCH1 mutations in chronic lymphocytic leukemia. Leukemia 2013; 28: 1060–1070.
Villamor N, Conde L, Martinez-Trillos A, Cazorla M, Navarro A, Bea S et al. NOTCH1 mutations identify a genetic subgroup of chronic lymphocytic leukemia patients with high risk of transformation and poor outcome. Leukemia 2013; 27: 1100–1106.
Weng AP, Ferrando AA, Lee W, Morris JP, Silverman LB, Sanchez-Irizarry C et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science 2004; 306: 269–271.
Wei P, Walls M, Qiu M, Ding R, Denlinger RH, Wong A et al. Evaluation of selective gamma-secretase inhibitor PF-03084014 for its antitumor efficacy and gastrointestinal safety to guide optimal clinical trial design. Mol Cancer Ther 2010; 9: 1618–1628.
Samon JB, Castillo-Martin M, Hadler M, mbesi-Impiobato A, Paietta E, Racevskis J et al. Preclinical analysis of the gamma-secretase inhibitor PF-03084014 in combination with glucocorticoids in T-cell acute lymphoblastic leukemia. Mol Cancer Ther 2012; 11: 1565–1575.
Ghia P, Stamatopoulos K, Belessi C, Moreno C, Stilgenbauer S, Stevenson F et al. ERIC recommendations on IGHV gene mutational status analysis in chronic lymphocytic leukemia. Leukemia 2007; 21: 1–3.
Rosich L, Saborit-Villarroya I, Lopez-Guerra M, Xargay-Torrent S, Montraveta A, Aymerich M et al. The phosphatidylinositol-3-kinase inhibitor NVP-BKM120 overcomes resistance signals derived from microenvironment by regulating the Akt/FoxO3a/Bim axis in chronic lymphocytic leukemia cells. Haematologica 2013; 98: 1739–1747.
Kim HS, Zhang X, Choi YS . Activation and proliferation of follicular dendritic cell-like cells by activated T lymphocytes. J Immunol 1994; 153: 2951–2961.
Xargay-Torrent S, Lopez-Guerra M, Saborit-Villarroya I, Rosich L, Campo E, Roue G et al. Vorinostat-induced apoptosis in mantle cell lymphoma is mediated by acetylation of proapoptotic BH3-only gene promoters. Clin Cancer Res 2011; 17: 3956–3968.
Palomero T, Lim WK, Odom DT, Sulis ML, Real PJ, Margolin A et al. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc Natl Acad Sci USA 2006; 103: 18261–18266.
Rosenwald A, Chuang EY, Davis RE, Wiestner A, Alizadeh AA, Arthur DC et al. Fludarabine treatment of patients with chronic lymphocytic leukemia induces a p53-dependent gene expression response. Blood 2004; 104: 1428–1434.
Espinosa L, Cathelin S, D'Altri T, Trimarchi T, Statnikov A, Guiu J et al. The Notch/Hes1 pathway sustains NF-kappaB activation through CYLD repression in T cell leukemia. Cancer Cell 2010; 18: 268–281.
Rosati E, Sabatini R, De FF, Del PB, Falzetti F, Di IM et al. Gamma-Secretase inhibitor I induces apoptosis in chronic lymphocytic leukemia cells by proteasome inhibition, endoplasmic reticulum stress increase and notch down-regulation. Int J Cancer 2013; 132: 1940–1953.
Landau DA, Carter SL, Stojanov P, McKenna A, Stevenson K, Lawrence MS et al. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell 2013; 152: 714–726.
Nakamura M, Shimada K, Konishi N . The role of HRK gene in human cancer. Oncogene 2008; 27 (Suppl 1): S105–S113.
Nakanishi C, Toi M . Nuclear factor-kappaB inhibitors as sensitizers to anticancer drugs. Nat Rev Cancer 2005; 5: 297–309.
Nwabo Kamdje AH, Bassi G, Pacelli L, Malpeli G, Amati E, Nichele I et al. Role of stromal cell-mediated Notch signaling in CLL resistance to chemotherapy. Blood Cancer J 2012; 2: e73.
Letilovic T, Vrhovac R, Verstovsek S, Jaksic B, Ferrajoli A . Role of angiogenesis in chronic lymphocytic leukemia. Cancer 2006; 107: 925–934.
Funahashi Y, Shawber CJ, Sharma A, Kanamaru E, Choi YK, Kitajewski J . Notch modulates VEGF action in endothelial cells by inducing matrix metalloprotease activity. Vasc Cell 2011; 3: 2.
Wang J, Fu L, Gu F, Ma Y . Notch1 is involved in migration and invasion of human breast cancer cells. Oncol Rep 2011; 26: 1295–1303.
Wang Z, Banerjee S, Li Y, Rahman KM, Zhang Y, Sarkar FH . Down-regulation of notch-1 inhibits invasion by inactivation of nuclear factor-kappaB, vascular endothelial growth factor, and matrix metalloproteinase-9 in pancreatic cancer cells. Cancer Res 2006; 66: 2778–2784.
Garg P, Jeppsson S, Dalmasso G, Ghaleb AM, McConnell BB, Yang VW et al. Notch1 regulates the effects of matrix metalloproteinase-9 on colitis-associated cancer in mice. Gastroenterology 2011; 141: 1381–1392.
Redondo-Munoz J, Ugarte-Berzal E, Terol MJ, Van den Steen PE, Hernandez del CM, Roderfeld M et al. Matrix metalloproteinase-9 promotes chronic lymphocytic leukemia b cell survival through its hemopexin domain. Cancer Cell 2010; 17: 160–172.
Kamiguti AS, Lee ES, Till KJ, Harris RJ, Glenn MA, Lin K et al. The role of matrix metalloproteinase 9 in the pathogenesis of chronic lymphocytic leukaemia. Br J Haematol 2004; 125: 128–140.
Buggins AG, Levi A, Gohil S, Fishlock K, Patten PE, Calle Y et al. Evidence for a macromolecular complex in poor prognosis CLL that contains CD38, CD49d, CD44 and MMP-9. Br J Haematol 2011; 154: 216–222.
Heinhuis B, Koenders MI, van den Berg WB, Netea MG, Dinarello CA, Joosten LA . Interleukin 32 (IL-32) contains a typical alpha-helix bundle structure that resembles focal adhesion targeting region of focal adhesion kinase-1. J Biol Chem 2012; 287: 5733–5743.
Sai J, Raman D, Liu Y, Wikswo J, Richmond A . Parallel phosphatidylinositol 3-kinase (PI3K)-dependent and Src-dependent pathways lead to CXCL8-mediated Rac2 activation and chemotaxis. J Biol Chem 2008; 283: 26538–26547.
Niggli V, Schlicht D, Affentranger S . Specific roles of Rac1 and Rac2 in motile functions of HT1080 fibrosarcoma cells. Biochem Biophys Res Commun 2009; 386: 688–692.
Acknowledgements
We thank Laura Jiménez and Sandra Cabezas for their technical support. We thank Dr Maria C Cid for providing HUVEC cells and Genomics Unit of IDIBAPS for the technical help. PF-03084014 was kindly provided by Pfizer. We are also very grateful to all individuals with CLL who have participated in this study from the CLL Spanish Consortium. This work was carried out at the Esther Koplowitz Center, Barcelona. This study was supported by research funding from Ministerio de Ciencia e Innovación (SAF 12/31242 to DC and SAF 11/29326 to PP-G), Fondo de Investigación Sanitaria (PI12/01847 to GR), Redes Temáticas de Investigación Cooperativa de Cáncer from the Instituto de Salud Carlos III (ISCIII), Spanish Ministry of Economy and Competitiveness and European Regional Development Fund (ERDF) ‘Una manera de hacer Europa’ RD12/0036/0004, RD12/0036/0036; RD12/0036/0023; RD12/0036/0067 and Generalitat de Catalunya 2009SGR967 (to DC). ML-G has a contract from Fundación Científica de la Asociación Española contra el Cáncer. AM and AM-C are recipients of predoctoral fellowships FPI from Ministerio de Ciencia e Innovación. PP-G holds a contract from Ramón y Cajal program (RYC2009-05134) and GR from Miguel Servet program (PI09/00060).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Leukemia website
Supplementary information
Rights and permissions
About this article
Cite this article
López-Guerra, M., Xargay-Torrent, S., Rosich, L. et al. The γ-secretase inhibitor PF-03084014 combined with fludarabine antagonizes migration, invasion and angiogenesis in NOTCH1-mutated CLL cells. Leukemia 29, 96–106 (2015). https://doi.org/10.1038/leu.2014.143
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2014.143
This article is cited by
-
High-resolution Melting Analysis for NOTCH1 c.7541-7542delCT Mutation in Chronic Lymphocytic Leukemia: Prognostic Significance in Egyptian Patients
Indian Journal of Hematology and Blood Transfusion (2022)
-
Proposal and validation of a method to classify genetic subtypes of diffuse large B cell lymphoma
Scientific Reports (2021)
-
Specific NOTCH1 antibody targets DLL4-induced proliferation, migration, and angiogenesis in NOTCH1-mutated CLL cells
Oncogene (2020)
-
Notch1 signaling in NOTCH1-mutated mantle cell lymphoma depends on Delta-Like ligand 4 and is a potential target for specific antibody therapy
Journal of Experimental & Clinical Cancer Research (2019)
-
Prolyl-isomerase Pin1 controls Notch3 protein expression and regulates T-ALL progression
Oncogene (2016)