Abstract
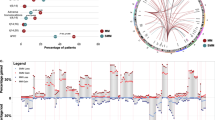
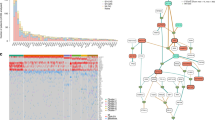
Although intratumor heterogeneity has been inferred in multiple myeloma (MM), little is known about its subclonal phylogeny. To describe such phylogenetic trees in a series of patients with MM, we perform whole-exome sequencing and single-cell genetic analysis. Our results demonstrate that at presentation myeloma is composed of two to six different major clones, which are related by linear and branching phylogenies. Remarkably, the earliest myeloma-initiating clones, some of which only had the initiating t(11;14), were still present at low frequencies at the time of diagnosis. For the first time in myeloma, we demonstrate parallel evolution whereby two independent clones activate the RAS/MAPK pathway through RAS mutations and give rise subsequently to distinct subclonal lineages. We also report the co-occurrence of RAS and interferon regulatory factor 4 (IRF4) p.K123R mutations in 4% of myeloma patients. Lastly, we describe the fluctuations of myeloma subclonal architecture in a patient analyzed at presentation and relapse and in NOD/SCID-IL2Rγnull xenografts, revealing clonal extinction and the emergence of new clones that acquire additional mutations. This study confirms that myeloma subclones exhibit different survival properties during treatment or mouse engraftment. We conclude that clonal diversity combined with varying selective pressures is the essential foundation for tumor progression and treatment resistance in myeloma.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Hanahan D, Weinberg RA . Hallmarks of cancer: the next generation. Cell 2011; 144: 646–674.
Nowell PC . The clonal evolution of tumor cell populations. Science 1976; 194: 23–28.
Greaves M, Maley CC . Clonal evolution in cancer. Nature 2012; 481: 306–313.
Anderson K, Lutz C, van Delft FW, Bateman CM, Guo Y, Colman SM et al. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature 2011; 469: 356–361.
Keats JJ, Chesi M, Egan JB, Garbitt VM, Palmer SE, Braggio E et al. Clonal competition with alternating dominance in multiple myeloma. Blood 2012; 120: 1067–1076.
Merlo LM, Pepper JW, Reid BJ, Maley CC . Cancer as an evolutionary and ecological process. Nat Rev Cancer 2006; 6: 924–935.
Sottoriva A, Spiteri I, Piccirillo SG, Touloumis A, Collins VP, Marioni JC et al. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci USA 2013; 110: 4009–4014.
Ding L, Ley TJ, Larson DE, Miller CA, Koboldt DC, Welch JS et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature 2012; 481: 506–510.
Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 2012; 366: 883–892.
Nik-Zainal S, Van Loo P, Wedge DC, Alexandrov LB, Greenman CD, Lau KW et al. The life history of 21 breast cancers. Cell 2012; 149: 994–1007.
Walker BA, Wardell CP, Melchor L, Hulkki S, Potter NE, Johnson DC et al. Intraclonal heterogeneity and distinct molecular mechanisms characterize the development of t(4;14) and t(11;14) myeloma. Blood 2012; 120: 1077–1086.
Campbell PJ, Yachida S, Mudie LJ, Stephens PJ, Pleasance ED, Stebbings LA et al. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature 2010; 467: 1109–1113.
Greaves M . Cancer stem cells as 'units of selection'. Evol Appl 2013; 6: 102–108.
Kuehl WM, Bergsagel PL . Molecular pathogenesis of multiple myeloma and its premalignant precursor. J Clin Invest 2012; 122: 3456–3463.
Morgan GJ, Walker BA, Davies FE . The genetic architecture of multiple myeloma. Nat Rev Cancer 2012; 12: 335–348.
Chapman MA, Lawrence MS, Keats JJ, Cibulskis K, Sougnez C, Schinzel AC et al. Initial genome sequencing and analysis of multiple myeloma. Nature 2011; 471: 467–472.
Egan JB, Shi CX, Tembe W, Christoforides A, Kurdoglu A, Sinari S et al. Whole-genome sequencing of multiple myeloma from diagnosis to plasma cell leukemia reveals genomic initiating events, evolution, and clonal tides. Blood 2012; 120: 1060–1066.
Magrangeas F, Avet-Loiseau H, Gouraud W, Lode L, Decaux O, Godmer P et al. Minor clone provides a reservoir for relapse in multiple myeloma. Leukemia 2013; 27: 473–481.
Durinck S, Ho C, Wang NJ, Liao W, Jakkula LR, Collisson EA et al. Temporal dissection of tumorigenesis in primary cancers. Cancer Discov 2011; 1: 137–143.
Potter NE, Ermini L, Papaemmanuil E, Cazzaniga G, Vijayaraghavan G, Titley I et al. Single-cell mutational profiling and clonal phylogeny in cancer. Genome Res 2013; 23: 2115–2125.
Boyd KD, Ross FM, Chiecchio L, Dagrada GP, Konn ZJ, Tapper WJ et al. A novel prognostic model in myeloma based on co-segregating adverse FISH lesions and the ISS: analysis of patients treated in the MRC Myeloma IX trial. Leukemia 2012; 26: 349–355.
Chiecchio L, Protheroe RK, Ibrahim AH, Cheung KL, Rudduck C, Dagrada GP et al. Deletion of chromosome 13 detected by conventional cytogenetics is a critical prognostic factor in myeloma. Leukemia 2006; 20: 1610–1617.
Walker BA, Wardell CP, Johnson DC, Kaiser MF, Begum DB, Dahir NB et al. Characterization of IGH locus breakpoints in multiple myeloma indicates a subset of translocations appear to occur in pregerminal center B cells. Blood 2013; 121: 3413–3419.
Walker BA, Leone PE, Chiecchio L, Dickens NJ, Jenner MW, Boyd KD et al. A compendium of myeloma-associated chromosomal copy number abnormalities and their prognostic value. Blood 2010; 116: e56–e65.
Walker BA, Leone PE, Jenner MW, Li C, Gonzalez D, Johnson DC et al. Integration of global SNP-based mapping and expression arrays reveals key regions, mechanisms, and genes important in the pathogenesis of multiple myeloma. Blood 2006; 108: 1733–1743.
Weaver S, Dube S, Mir A, Qin J, Sun G, Ramakrishnan R et al. Taking qPCR to a higher level: analysis of CNV reveals the power of high throughput qPCR to enhance quantitative resolution. Methods 2010; 50: 271–276.
Sims D, Bursteinas B, Gao Q, Jain E, MacKay A, Mitsopoulos C et al. ROCK: a breast cancer functional genomics resource. Breast Cancer Res Treat 2010; 124: 567–572.
Fryer RA, Graham TJ, Smith EM, Walker-Samuel S, Morgan GJ, Robinson SP et al. Characterization of a novel mouse model of multiple myeloma and its use in preclinical therapeutic assessment. PLoS One 2013; 8: e57641.
Scrucca L . GA: A package for genetic algorithms in R. J Stat Softw 2013; 53: 1–37.
Fonseca R, Bailey RJ, Ahmann GJ, Rajkumar SV, Hoyer JD, Lust JA et al. Genomic abnormalities in monoclonal gammopathy of undetermined significance. Blood 2002; 100: 1417–1424.
Walker BA, Wardell CP, Melchor L, Brioli A, Johnson DC, Kaiser MF et al. Intraclonal heterogeneity is a critical early event in the development of myeloma and precedes the development of clinical symptoms. Leukemia 2013; e-pub ahead of print 2 July 2013 doi:10.1038/leu.2013.199.
Park SY, Gonen M, Kim HJ, Michor F, Polyak K . Cellular and genetic diversity in the progression of in situ human breast carcinomas to an invasive phenotype. J Clin Invest 2010; 120: 636–644.
Newburger DE, Kashef-Haghighi D, Weng Z, Salari R, Sweeney RT, Brunner AL et al. Genome evolution during progression to breast cancer. Genome Res 2013; 23: 1097–1108.
Merlo LM, Shah NA, Li X, Blount PL, Vaughan TL, Reid BJ et al. A comprehensive survey of clonal diversity measures in Barrett's esophagus as biomarkers of progression to esophageal adenocarcinoma. Cancer Prev Res (Phila) 2010; 3: 1388–1397.
Forrester K, Almoguera C, Han K, Grizzle WE, Perucho M . Detection of high incidence of K-ras oncogenes during human colon tumorigenesis. Nature 1987; 327: 298–303.
Farr CJ, Saiki RK, Erlich HA, McCormick F, Marshall CJ . Analysis of RAS gene mutations in acute myeloid leukemia by polymerase chain reaction and oligonucleotide probes. Proc Natl Acad Sci USA 1988; 85: 1629–1633.
Itzykson R, Kosmider O, Renneville A, Morabito M, Preudhomme C, Berthon C et al. Clonal architecture of chronic myelomonocytic leukemias. Blood 2013; 121: 2186–2198.
Andrulis M, Lehners N, Capper D, Penzel R, Heining C, Huellein J et al. Targeting the BRAF V600E mutation in multiple myeloma. Cancer Discov 2013; 3: 862–869.
Lu R . Interferon regulatory factor 4 and 8 in B-cell development. Trends Immunol 2008; 29: 487–492.
Lamy L, Ngo VN, Emre NC, Shaffer AL 3rd, Yang Y, Tian E et al. Control of autophagic cell death by caspase-10 in multiple myeloma. Cancer Cell 2013; 23: 435–449.
Shaffer AL, Emre NC, Lamy L, Ngo VN, Wright G, Xiao W et al. IRF4 addiction in multiple myeloma. Nature 2008; 454: 226–231.
Lopez-Girona A, Mendy D, Ito T, Miller K, Gandhi AK, Kang J et al. Cereblon is a direct protein target for immunomodulatory and antiproliferative activities of lenalidomide and pomalidomide. Leukemia 2012; 26: 2326–2335.
Zhang LH, Kosek J, Wang M, Heise C, Schafer PH, Chopra R . Lenalidomide efficacy in activated B-cell-like subtype diffuse large B-cell lymphoma is dependent upon IRF4 and cereblon expression. Br J Haematol 2013; 160: 487–502.
Zhu YX, Braggio E, Shi CX, Bruins LA, Schmidt JE, Van Wier S et al. Cereblon expression is required for the antimyeloma activity of lenalidomide and pomalidomide. Blood 2011; 118: 4771–4779.
Havelange V, Pekarsky Y, Nakamura T, Palamarchuk A, Alder H, Rassenti L et al. IRF4 mutations in chronic lymphocytic leukemia. Blood 2011; 118: 2827–2829.
Swanton C . Intratumor heterogeneity: evolution through space and time. Cancer Res 2012; 72: 4875–4882.
Acknowledgements
We acknowledge Professor Mel Greaves for his scientific input and the Tumour Profiling Unit at the Institute of Cancer Research for their support and technical expertise in this study. This study was supported by a Cancer Research UK sample collection grant (CTAAC), grants from the National Institutes of Health Biomedical Research Centre at the Royal Marsden Hospital and a Myeloma UK programme grant (LM, CPW, RAF, DCJ and BAW). MFK is supported by a Research Fellowship by Deutsche Forschungsgemeinschaft (KA 3338/1-1). FED is a Cancer Research UK Senior Clinical Fellow.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Leukemia website
Supplementary information
Rights and permissions
About this article
Cite this article
Melchor, L., Brioli, A., Wardell, C. et al. Single-cell genetic analysis reveals the composition of initiating clones and phylogenetic patterns of branching and parallel evolution in myeloma. Leukemia 28, 1705–1715 (2014). https://doi.org/10.1038/leu.2014.13
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2014.13
Keywords
This article is cited by
-
Bispecific CS1-BCMA CAR-T cells are clinically active in relapsed or refractory multiple myeloma
Leukemia (2024)
-
Mass Cytometry reveals unique phenotypic patterns associated with subclonal diversity and outcomes in multiple myeloma
Blood Cancer Journal (2023)
-
Correlation of plasma cell assessment by phenotypic methods and molecular profiles by NGS in patients with plasma cell dyscrasias
BMC Medical Genomics (2022)
-
Single-cell profiling of tumour evolution in multiple myeloma — opportunities for precision medicine
Nature Reviews Clinical Oncology (2022)
-
Bringing precision oncology to cellular resolution with single-cell genomics
Clinical & Experimental Metastasis (2022)