Abstract
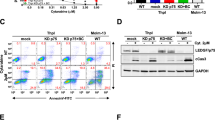
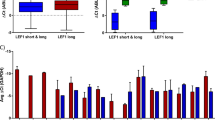
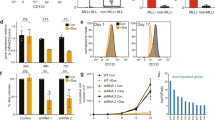
Mixed lineage leukemia (MLL) fusion proteins directly activate the expression of key downstream genes such as MEIS1, HOXA9 to drive an aggressive form of human leukemia. However, it is still poorly understood what additional transcriptional regulators, independent of the MLL fusion pathway, contribute to the development of MLL leukemia. Here we show that the transcription factor PU.1 is essential for MLL leukemia and is required for the growth of MLL leukemic cells via the promotion of cell-cycle progression and inhibition of apoptosis. Importantly, PU.1 expression is not under the control of MLL fusion proteins. We further identified a PU.1-governed 15-gene signature, which contains key regulators in the MEIS-HOX program (MEIS1, PBX3, FLT3, and c-KIT). PU.1 directly binds to the genomic loci of its target genes in vivo, and is required to maintain active expression of those genes in both normal hematopoietic stem and progenitor cells and in MLL leukemia. Finally, the clinical significance of the identified PU.1 signature was indicated by its ability to predict survival in acute myelogenous leukemia patients. Together, our findings demonstrate that PU.1 contributes to the development of MLL leukemia, partially via crosstalk with the MEIS/HOX pathway.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Krivtsov A, Armstrong S . MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer 2007; 7: 823.
de Boer J, Walf-Vorderwülbecke V, Williams O . In focus: MLL rearranged leukemia. Leukemia 2013; 27: 1224–1228.
Meyer C, Kowarz E, Hofmann J, Renneville A, Zuna J, Trka J et al. New insights to the MLL recombinome of acute leukemias. Leukemia 2009; 23: 1490–1499.
Nakamura T, Alder H, Gu Y, Prasad R, Canaani O, Kamada N et al. Genes on chromosomes 4, 9 and 19 involved in 11q23 abnormalities in acute leukemia share sequence homology and/or common motifs. Natl Acad Sci U S A 1993; 90: 4631–4635.
Bitoun E, Oliver P, Davies K . The mixed-lineage leukemia fusion partner AF4 stimulates RNA polymerase II transcriptional elongation and mediates coordinated chromatin remodeling. Hum Mol Genet 2007; 16: 92.
Mueller D, Bach C, Zeisig D, Garcia-Cuellar M, Monroe S, Sreekumar A et al. A role for the MLL fusion partner ENL in transcriptional elongation and chromatin modification. Blood 2007; 110: 4445.
Greaves M, Wiemels J . Origins of chromosome translocations in childhood leukaemia. Nat Rev Cancer 2003; 3: 639.
Balgobind B, Zwaan C, Pieters R, Van den Heuvel-Eibrink M . The heterogeneity of pediatric MLL-rearranged acute myeloid leukemia. Leukemia 2011; 25: 1239–1248.
Rosenbauer F, Tenen DG . Transcription factors in myeloid development: balancing differentiation with transformation. Nat Rev Immunol 2007; 7: 105–117.
Armstrong S, Staunton J, Silverman L, Pieters R, den Boer M, Minden M et al. MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat Genet 2002; 30: 41.
Valk PJM, Verhaak RGW, Beijen MA, Erpelinck CAJ, van Doorn-Khosrovani SBW, Boer JM et al. Prognostically useful gene-expression profiles in acute myeloid leukemia. N Engl J Med 2004; 350: 1617–1628.
Ross ME, Mahfouz R, Onciu M, Liu HC, Zhou X, Song G et al. Gene expression profiling of pediatric acute myelogenous leukemia. Blood 2004; 104: 3679–3687.
Zeisig BB, Milne T, García-Cuéllar MP, Schreiner S, Martin ME, Fuchs U et al. Hoxa9 and Meis1 are key targets for MLL-ENL-mediated cellular immortalization. Mol Cell Biol 2004; 24: 617–628.
Martin ME, Milne TA, Bloyer S, Galoian K, Shen W, Gibbs D et al. Dimerization of MLL fusion proteins immortalizes hematopoietic cells. Cancer cell 2003; 4: 197–207.
So CW, Lin M, Ayton PM, Chen EH, Cleary ML . Dimerization contributes to oncogenic activation of MLL chimeras in acute leukemias. Cancer cell 2003; 4: 99–110.
Ayton PM, Cleary ML . Transformation of myeloid progenitors by MLL oncoproteins is dependent on Hoxa7 and Hoxa9. Genes Dev 2003; 17: 2298.
Wang J, Iwasaki H, Krivtsov A, Febbo PG, Thorner AR, Ernst P et al. Conditional MLL-CBP targets GMP and models therapy-related myeloproliferative disease. EMBO J 2005; 24: 368.
Okada Y, Feng Q, Lin Y, Jiang Q, Li Y, Coffield V et al. hDOT1L links histone methylation to leukemogenesis. Cell 2005; 121: 167.
Kumar A, Hudson W, Chen W, Nishiuchi R, Yao Q, Kersey J . Hoxa9 influences the phenotype but not the incidence of Mll-AF9 fusion gene leukemia. Blood 2004; 103: 1823.
Kumar AR, Li Q, Hudson WA, Chen W, Sam T, Yao Q et al. A role for MEIS1 in MLL-fusion gene leukemia. Blood 2009; 113: 1756.
Wong P, Iwasaki M, Somervaille TC, So CWE, Cleary ML . Meis1 is an essential and rate-limiting regulator of MLL leukemia stem cell potential. Genes Dev 2007; 21: 2762.
Mann RS, Lelli KM, Joshi R . Hox specificity: unique roles for cofactors and collaborators. Curr Top Dev Biol 2009; 88: 63–101.
Huang Y, Sitwala K, Bronstein J, Sanders D, Dandekar M, Collins C et al. Identification and characterization of Hoxa9 binding sites in hematopoietic cells. Blood 2012; 119: 388–398.
Hu Y-L, Fong S, Ferrell C, Largman C, Shen W-F . HOXA9 modulates its oncogenic partner Meis1 to influence normal hematopoiesis. Mol Cell Biol 2009; 29: 5181–5192.
Wang GG, Pasillas MP, Kamps MP . Persistent transactivation by Meis1 replaces Hox function in myeloid leukemogenesis models: evidence for co-occupancy of Meis1-Pbx and Hox-Pbx complexes on promoters of leukemia-associated genes. Mol Cell Biol 2006; 26: 3902.
Wang GG, Pasillas MP, Kamps MP . Meis1 programs transcription of FLT3 and cancer stem cell character, using a mechanism that requires interaction with Pbx and a novel function of the Meis1 C-terminus. Blood 2005; 106: 254–264.
Wang Q-f, Li Y-j, Dong J-f, Li B, Kaberlein JJ, Zhang L et al. Regulation of MEIS1 by distal enhancer elements in acute leukemia. Leukemia 2013; e-pub ahead of print 11 September 2013; doi:10.1038/leu.2013.260.
Huang G, Zhang P, Hirai H, Elf S, Yan X, Chen Z et al. PU. 1 is a major downstream target of AML1 (RUNX1) in adult mouse hematopoiesis. Nature Genet 2008; 40: 51.
Iwasaki H, Somoza C, Shigematsu H, Duprez EA, Iwasaki-Arai J, Mizuno S et al. Distinctive and indispensable roles of PU. 1 in maintenance of hematopoietic stem cells and their differentiation. Blood 2005; 106: 1590–1600.
Rosenbauer F, Wagner K, Kutok JL, Iwasaki H, Le Beau MM, Okuno Y et al. Acute myeloid leukemia induced by graded reduction of a lineage-specific transcription factor, PU. 1. Nature Genet 2004; 36: 624–630.
Zhang Y, Yan X, Sashida G, Zhao X, Rao Y, Goyama S et al. Stress hematopoiesis reveals abnormal control of self-renewal, lineage bias, and myeloid differentiation in Mll partial tandem duplication (Mll-PTD) hematopoietic stem/progenitor cells. Blood 2012; 120: 1118–1129.
Verhaak RG, Wouters BJ, Erpelinck CA, Abbas S, Beverloo HB, Lugthart S et al. Prediction of molecular subtypes in acute myeloid leukemia based on gene expression profiling. Haematologica 2009; 94: 131.
Tomasson MH, Xiang Z, Walgren R, Zhao Y, Kasai Y, Miner T et al. Somatic mutations and germline sequence variants in the expressed tyrosine kinase genes of patients with de novo acute myeloid leukemia. Blood 2008; 111: 4797.
Haferlach T, Kohlmann A, Wieczorek L, Basso G, Te Kronnie G, Béné M-C et al. Clinical utility of microarray-based gene expression profiling in the diagnosis and subclassification of leukemia: report from the International Microarray Innovations in Leukemia Study Group. J Clin Oncol 2010; 28: 2529–2537.
Metzeler KH, Hummel M, Bloomfield CD, Spiekermann K, Braess J, Sauerland MC et al. An 86-probe-set gene-expression signature predicts survival in cytogenetically normal acute myeloid leukemia. Blood 2008; 112: 4193–4201.
Wilson NK, Foster SD, Wang X, Knezevic K, Schütte J, Kaimakis P et al. Combinatorial transcriptional control in blood stem/progenitor cells: genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell 2010; 7: 532–544.
Lefterova MI, Steger DJ, Zhuo D, Qatanani M, Mullican SE, Tuteja G et al. Cell-Specific Determinants of Peroxisome Proliferator-Activated Receptor γ Function in Adipocytes and Macrophages. Mol Cell Biol 2010; 30: 2078.
Barash Y, Dehan E, Krupsky M, Franklin W, Geraci M, Friedman N et al. Comparative analysis of algorithms for signal quantitation from oligonucleotide microarrays. Bioinformatics 2004; 20: 839.
Wang Q-f, Wu G, Mi S, He F, Wu J, Dong J et al. MLL fusion proteins preferentially regulate a subset of wild-type MLL target genes in the leukemic genome. Blood 2011; 117: 6895–6905.
Bernt KM, Zhu N, Sinha AU, Vempati S, Faber J, Krivtsov AV et al. MLL-Rearranged Leukemia Is Dependent on Aberrant H3K79 Methylation by DOT1L. Cancer cell 2011; 20: 66–78.
Zuber J, Rappaport AR, Luo W, Wang E, Chen C, Vaseva AV et al. An integrated approach to dissecting oncogene addiction implicates a Myb-coordinated self-renewal program as essential for leukemia maintenance. Genes Dev 2011; 25: 1628–1640.
Guenther MG, Lawton LN, Rozovskaia T, Frampton GM, Levine SS, Volkert TL et al. Aberrant chromatin at genes encoding stem cell regulators in human mixed-lineage leukemia. Genes Dev 2008; 22: 3403–3408.
Knezevic K, Bee T, Wilson NK, Janes ME, Kinston S, Polderdijk S et al. A Runx1-Smad6 Rheostat Controls Runx1 Activity during Embryonic Hematopoiesis. Mol Cell Biol 2011; 31: 2817.
Poirel H, Rack K, Delabesse E, Radford-Weiss I, Troussard X, Debert C et al. Incidence and characterization of MLL gene (11q23) rearrangements in acute myeloid leukemia M1 and M5. Blood 1996; 87: 2496–2505.
Rabbitts T . Chromosomal translocations in human cancer. Nature 1994; 372: 143.
Swansbury G . The proportion of clonal divisions varies in different hematologic malignancies. The United Kingdom Cancer Cytogenetics Group (UKCCG)[corrected]. Cancer Genet Cytogenet 1998; 104: 139.
Vangala RK, Heiss-Neumann MS, Rangatia JS, Singh SM, Schoch C, Tenen DG et al. The myeloid master regulator transcription factor PU. 1 is inactivated by AML1-ETO in t (8; 21) myeloid leukemia. Blood 2003; 101: 270–277.
Wang K, Wang P, Shi J, Zhu X, He M, Jia X et al. PML/RARalpha targets promoter regions containing PU. 1 consensus and RARE half sites in acute promyelocytic leukemia. Cancer cell 2010; 17: 186.
Mueller BU, Pabst T, Fos J, Petkovic V, Fey MF, Asou N et al. ATRA resolves the differentiation block in t (15; 17) acute myeloid leukemia by restoring PU. 1 expression. Blood 2006; 107: 3330.
Harrell F, Lee KL, Mark DB . Tutorial in biostatistics multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat Med 1996; 15: 361–387.
DerSimonian R, Laird N . Meta-analysis in clinical trials. Control Clin Trials 1986; 7: 177–188.
Venet D, Dumont JE, Detours V . Most random gene expression signatures are significantly associated with breast cancer outcome. PLoS Comput Biol 2011; 7: e1002240.
Cook WD, McCaw BJ, Herring C, John DL, Foote SJ, Nutt SL et al. PU. 1 is a suppressor of myeloid leukemia, inactivated in mice by gene deletion and mutation of its DNA binding domain. Blood 2004; 104: 3437–3444.
Mueller BU, Pabst T, Osato M, Asou N, Johansen LM, Minden MD et al. Heterozygous PU. 1 mutations are associated with acute myeloid leukemia. Blood 2002; 100: 998–1007.
Aikawa Y, Katsumoto T, Zhang P, Shima H, Shino M, Terui K et al. PU. 1-mediated upregulation of CSF1R is crucial for leukemia stem cell potential induced by MOZ-TIF2. Nature Med 2010; 16: 580–585.
Staber P, Zhang P, Ye M, Welner R, Nombela-Arrieta C, Bach C et al. Sustained PU. 1 levels balance cell-cycle regulators to prevent exhaustion of adult hematopoietic stem cells. Mol Cell 2013; 49: 934–946.
Acknowledgements
We thank Drs Jose Cancelas, H Leighton Grimes, James Mulloy, Susumu Goyama (Cincinnati Children’s Hospital Medical Center) and Frank Rosenbauer (Max Delbrück Center for Molecular Medicine) for their critical reading and valuable comments on the manuscript. We also thank Tami R Bartell for English editing. This study was supported by the Hundred Talents Program of the Chinese Academy of Sciences (to QFW), the National Natural Science Foundation of China grants No. 81070442 and No. 91331111 (to QFW), No. 81100381 (to JZ), No. 81300438 (to JW) and the ‘Strategic Priority Research Program’ of the Chinese Academy of Sciences (to QFW), No. XDA01010305. This work was also supported in part by the Knowledge Innovation Program of the Chinese Academy of Sciences (to FHH) and sponsored by the Scientific Research Foundation for the Returned Overseas Chinese Scholars, State Education Ministry (to QFW). We would like to acknowledge support from the Cincinnati Children’s Hospital Research Foundation, the Ohio Cancer Research Associates, the Cancer Free Kids, the Leukemia Research Foundation and the Pilot Research Grant of the State Key Laboratory of Experimental Hematology (Tianjin, China) (to GH), the NIH grant HL112719 (to DGT) and NIH grant HL111192 (to AK).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Leukemia website
Supplementary information
Rights and permissions
About this article
Cite this article
Zhou, J., Wu, J., Li, B. et al. PU.1 is essential for MLL leukemia partially via crosstalk with the MEIS/HOX pathway. Leukemia 28, 1436–1448 (2014). https://doi.org/10.1038/leu.2013.384
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2013.384
Keywords
This article is cited by
-
Mutational synergy during leukemia induction remodels chromatin accessibility, histone modifications and three-dimensional DNA topology to alter gene expression
Nature Genetics (2021)
-
LSD1 inhibition by tranylcypromine derivatives interferes with GFI1-mediated repression of PU.1 target genes and induces differentiation in AML
Leukemia (2019)
-
ANP32A regulates histone H3 acetylation and promotes leukemogenesis
Leukemia (2018)
-
Vitamin C-induced epigenomic remodelling in IDH1 mutant acute myeloid leukaemia
Leukemia (2018)
-
Recurrent SPI1 (PU.1) fusions in high-risk pediatric T cell acute lymphoblastic leukemia
Nature Genetics (2017)