Abstract
Reliable detection of JAK2-V617F is critical for accurate diagnosis of myeloproliferative neoplasms (MPNs); in addition, sensitive mutation-specific assays can be applied to monitor disease response. However, there has been no consistent approach to JAK2-V617F detection, with assays varying markedly in performance, affecting clinical utility. Therefore, we established a network of 12 laboratories from seven countries to systematically evaluate nine different DNA-based quantitative PCR (qPCR) assays, including those in widespread clinical use. Seven quality control rounds involving over 21 500 qPCR reactions were undertaken using centrally distributed cell line dilutions and plasmid controls. The two best-performing assays were tested on normal blood samples (n=100) to evaluate assay specificity, followed by analysis of serial samples from 28 patients transplanted for JAK2-V617F-positive disease. The most sensitive assay, which performed consistently across a range of qPCR platforms, predicted outcome following transplant, with the mutant allele detected a median of 22 weeks (range 6–85 weeks) before relapse. Four of seven patients achieved molecular remission following donor lymphocyte infusion, indicative of a graft vs MPN effect. This study has established a robust, reliable assay for sensitive JAK2-V617F detection, suitable for assessing response in clinical trials, predicting outcome and guiding management of patients undergoing allogeneic transplant.
Similar content being viewed by others
Introduction
The discovery of the JAK2-V617F mutation in 2005 transformed the diagnostic work-up of myeloproliferative neoplasms (MPNs), which occurs in ∼98% of polycythemia vera (PV) cases and ∼50% with primary myelofibrosis (PMF) and essential thrombocythemia (ET).1, 2, 3, 4, 5, 6 It has become clear that the level of the mutant allele detectable in bona fide MPN cases can be as low as 1–3%,6, 7, 8, 9 which has implications for the diagnostic methods used, as such a level would not be detectable by conventional Sanger sequencing or pyrosequencing, and could potentially be missed by allele-specific/amplification refractory mutation system PCR.10, 11, 12 Quantitative PCR (qPCR) can afford much greater specificity and thereby sensitivity; a considerable number of assays have been developed,10 which have been applied to quantify the relative level of the mutant allele, to gain further insights into disease biology and distinguish subsets of patients with differing clinical features and outcome. Mutant allele burden has been correlated with the risk of thrombotic events in ET, severity of the disease phenotype in PV and survival in PMF.13, 14, 15, 16, 17, 18, 19, 20 JAK2-V617F has also been quantified to assess disease response, with significant reductions in allele burden reported in PV patients following interferon α-2b and pegylated interferon α-2a treatment.21, 22, 23, 24, 25 Moreover, serial DNA-based qPCR assays have been used after allogeneic transplantation for PMF to predict outcome,26, 27, 28, 29, 30 with patients with an allele burden >1% on day 28 post-transplant30 or failing to achieve molecular remission in the peripheral blood (PB) by 6 months post-allograft being at a significant risk of relapse,29 which can potentially be prevented by donor lymphocyte infusion (DLI).26, 31, 32
In addition to helping guide management in the transplant setting, quantification of JAK2-V617F is being considered as an end point in a number of trials with novel agents, including JAK inhibitors. Studies to date indicate that JAK inhibitors have a relatively limited impact on JAK2-V617F allele burden,33, 34, 35, 36 whereas, a recent study with the novel telomerase inhibitor imetelstat, showed a substantial decrease in JAK2-V617F allele burden.37 It is critically important that assays used at baseline to potentially distinguish biological subsets of disease and subsequently to track treatment response are robust, mutant-specific and afford a suitable level of specificity. On the basis of experience of molecular monitoring in other hematological malignancies within the European LeukemiaNet (ELN), we reasoned that the plethora of qPCR assays routinely used to detect JAK2-V617F could vary markedly in their performance, potentially impacting on their clinical utility. Therefore, we established a joint initiative between working groups within the ELN and MPN&MPNr-EuroNet to evaluate in systematic fashion a large number of widely used published and ‘in-house’ JAK2-V617F qPCR assays, leading to the identification of an assay that, beyond being robust enough for routine diagnostic purposes, also showed the best performance profile when used for predicting outcome following an allogeneic transplant.
Materials and methods
Detection of JAK2-V617F mutant allele by quantitative PCR (qPCR) assays
Genomic DNA was extracted from cell lines and human PB samples using standard methods. DNA was amplified (25 ng template per well) using different qPCR assays designed to detect wild-type JAK2 (JAK2-WT) and the JAK2 1849G>T mutation encoding JAK2-V617F (designated JAK2-V617F assay), in parallel with independent control gene assays for Cyclophilin A38 and Albumin (ALB),39 providing a control for any variation in the amount of template DNA between reaction wells, as well as between successive quality control (QC) rounds (see Supplementary Table 1 for primer and probe sequences). The evaluation encompassed a number of unpublished ‘in-house’ assays (n=3), together with a large range of published assays (n=6).26, 40, 41, 42, 43, 44 The latter included assays that have been widely applied in clinical practice, that is, those forming the basis of the MutaQuant kit (Qiagen, Marseille, France),41 or reported to predict outcome and used to guide management following allogeneic transplantation.26, 29, 31, 32 For published assays, reported reaction conditions were used; whereas for ‘in-house’ assays, the reaction conditions used were provided by the respective source laboratories. Assays were run in duplicate in the QC rounds and in triplicate wells for the analysis of control PB and primary patient samples with appropriate water controls.
Selection of the best-performing JAK2-V617F qPCR assay
A network of 12 laboratories with a particular interest in molecular diagnosis of myeloid neoplasms and development of real-time qPCR assays for minimal residual disease (MRD) detection was established by the ELN in conjunction with the MPN&MPNr-EuroNet consortium (see www.leukemia-net.org/ and www.mpneuronet.eu/). The study involved central distribution of test materials for seven QC rounds comprising cell line DNA and plasmid standards (see Figure 1). qPCR reagents were also centrally distributed together with reaction conditions to be used for each respective assay, which was assigned a different code number in each successive QC round. qPCRs were predominantly performed on Applied Biosystems platforms (ABI7000/7300/7500/7900; Applied Biosystems, Foster City, CA, USA), two laboratories used the Lightcycler LC480 platform (Roche Applied Science, Penzberg, Germany) and one used a Rotor-Gene 6000 instrument (Corbett Life Science (now Qiagen), Sydney, Australia). Laboratories with established published or in-house qPCR assays were encouraged to perform these in parallel with the coded test assays on the distributed QC materials, to control for local variations in test conditions. Raw data from each QC round were submitted by participating laboratories for independent central analysis (Department of Genetics, King’s College, London); conclusions were drawn concerning the performance of the various assays before breaking the code to reveal their identities.
Summary of QC rounds used to identify JAK2-V617F assays with the best performance profile. Seven successive QC rounds, designated QC1–7, were conducted involving the assessment of plasmid standards and DNA derived from dilutions of cell lines harboring JAK2-V617F (HEL or UKE-1) in cells with wild-type JAK2 (K562 or PB mononuclear cells), abbreviated: HEL/K562, UKE-1/PBMNC, as indicated. Assays found to exhibit consistently poorer performance in multiple test laboratories were not taken forward to subsequent QC rounds—reasons for assay exclusion are provided. Details of the JAK2-V617F assays (designated Assays 1–9) and control gene assays (Albumin, Cyclophilin) are provided in Supplementary Table 1.
The relative efficiency of JAK2-WT, JAK2-V617F, Cyclophilin and ALB assays was determined using serial dilutions of respective plasmid standards (gift from Ipsogen (now Qiagen Marseille), Marseille, France), as described.45, 46 To investigate the specificity of the JAK2-WT and JAK2-V617F assays, genomic DNA extracted from undiluted HEL and K562 cells (which harbor only JAK2-V617F and wild-type JAK2, respectively) was analyzed. The capacity afforded by the different JAK2-V617F assays to detect the mutant allele was established through the analysis of DNA extracted from serial dilutions of JAK2-V617F mutant cells (HEL or UKE-1) in K562 cells. The level of JAK2-V617F at each dilution within the 5–100% range of the distributed cell line sample DNA was verified by pyrosequencing. The detection limit of each JAK2-V617F assay was designated as the lowest dilution of the mutant cell line giving rise to amplification significantly above the background level (that is, normalized JAK2-V617F ΔCt⩾1) observed in undiluted K562 cells and with a cycle threshold (Ct) value ⩽40. The specificity of the best-performing JAK2-V617F assays was investigated further, analyzing a large cohort of control PB samples (n=100) taken from individuals with no history of MPN. Overall, Assay 5 was found to have the best performance profile and was selected as the ELN/COST JAK2 assay: forward primer 5′-CTTTCTTTGAAGCAGCAAGTATGA-3′, reverse primer JAK2-WT 5′-GTAGTTTTACTTACTCTCGTCTCCACATAC-3′, reverse primer JAK2-V617F 5′-GTAGTTTTACTTACTCTCGTCTCCACATAA-3′, probe 6FAM-TGAGCAAGCTTTCTCACAAGCATTTGGTTTTAMRA.
Evaluation of JAK2-V617F qPCR assays in patients undergoing allogeneic transplantation
Stored PB DNA samples (n=220) from a cohort of 28 patients undergoing allogeneic transplantation for JAK2-V617F-associated myeloid neoplasms (2 PV, 16 myelofibrosis, 8 transformed MPN and 2 JAK2-V617F mutant chronic myelomonocytic leukemia) were analyzed. Given their older age (median 54 years, range 21–67 years), the majority received reduced intensity conditioning regimens. A median of seven samples were available per patient (range 3–14). All samples were considered evaluable on the basis of ALB Ct value (median 26, range 21–30). JAK2-V617F was quantified using the ΔCt method, as described.47 Samples were defined as qPCR positive for JAK2-V617F when the ΔCt value between the ALB and JAK2 mutant assays was below the background level of amplification detected in normal PB samples (see below) in at least two of three replicate wells with Ct values ⩽40 (threshold 0.1),that is, ΔCtJAK2-V617F-ALB <13.3 for Assay 5 and <11.7 for Assay 7. qPCR results were compared with chimerism data provided by the laboratories serving the respective transplant centers. Patient samples were provided following informed consent in accordance with the Declaration of Helsinki and analyses were subject to institutional ethical approval (Guy’s and St Thomas’ Local Research Ethics Committee ref 06/Q0702/140).
Results
QC rounds to identify JAK2-V617F qPCR assays with the best performance profile in the diagnostic (JAK2-V617F range: 0.5–100%) and MRD detection (JAK2-V617F <1%) ranges.
In order to obtain some information concerning possible variability in JAK2-V617F quantification between laboratories, freeze-dried cells from a 1 in 30 dilution of HEL cells (which harbor multiple copies of JAK2-V617F and lack the wild-type JAK2 allele) in K562 cells (WT JAK2) were provided to five ELN member laboratories by the UK National External Quality Assessment Service (UK NEQAS, Sheffield, UK). The test sample was estimated to comprise ∼10% JAK2-V617F; however, the recipient laboratories who each used their own established JAK2-V617F qPCR assay reported markedly differing levels of mutant allele percentage, ranging from 23 to 80% (see Supplementary Figure 1). This highlighted the limitations of categorization of MPNs as having heterozygous or homozygous JAK2-V617F mutations on the basis of conventional qPCR assays and emphasized the need for greater standardization. The aim was to identify qPCR assays with sufficient specificity and sensitivity, and comparable interlaboratory performance irrespective of qPCR platform, that are robust enough to be used for routine diagnostic purposes, as well as for tracking MRD after disease-modifying therapies (for example, interferon, stem cell transplantation), where a <1% detection limit is required. A series of seven QC rounds (QC1–7) were conducted (see Figure 1), which between them involved 12 laboratories (11 European, 1 US), spread between 7 countries. This analysis included consideration of a number of qPCR assays that have been widely reported to track treatment response, including post-transplant surveillance. In QC1, six assays were evaluated on DNA from the serial dilutions of HEL in K562 cells; qPCRs that used differently labeled probes to detect the mutant and wild-type alleles in the same reaction (that is, Assays 1(ref. 40) and 3(ref. 42)) were found to be too insensitive to be suitable for MRD detection and were not considered further. In addition, Assay 4(ref. 26) which has been widely applied for post-transplant surveillance, was observed to perform inconsistently between laboratories, and as it afforded poorer sensitivity compared with Assay 5 in head to head testing, it was also eliminated at this stage.
Assays 2(ref. 41), 5(ref. 43) and 6(ref. 44) were reanalyzed in QC2 and taken forward to QC3 which involved the evaluation of an additional in-house JAK2-V617F assay (Assay 7, Elisabeth Oppliger Leibundgut, University of Bern, unpublished), together with established assays for independent-control genes, that is, Cyclophilin A38 and ALB,39 which were tested on cell line dilutions, as well as plasmid standards (Figure 2). In contrast to the initial experience with analysis of the UK NEQAS test sample, the QC rounds showed a high level of concordance between the results obtained by the respective testing laboratories despite the use of different qPCR platforms, with plots of Ct values for each of the dilutions lying largely in parallel (Figure 3). Moreover, Assays 2, 5 and 7 were confirmed to provide reliable quantification between laboratories within the diagnostic range, as shown by close correlation with the percentage of JAK2-V617F, as determined by pyrosequencing (Supplementary Figure 2), indicating their suitability for use in the routine diagnostic setting.
Plasmid standards. In QC rounds QC3–7, in addition to the assessment on cell line dilutions, JAK2 and control gene assays were tested on serial dilutions of plasmid standards. The efficiencies of JAK2-V617F Assays 2, 5 and 7 were found to be superior to that of Assay 6, with slope values much closer to −3.32 (the slope associated with a PCR reaction with maximal efficiency). y-axis: Mean cycle threshold (Ct) values; x-axis: log dilution.
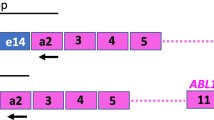
Interlaboratory concordance of JAK2-V617F quantification. Left panel: shows the design of Assays 2, 5 and 7, which were found to afford the greatest sensitivity for the detection of JAK2-V617F. Right panel: graphs show mean Ct value for the respective JAK2-V617F assays reported by each laboratory obtained on the analysis of DNA from serial dilutions of HEL in K562 cells in the third QC round (QC3). Plots of the dilution series were observed to lie largely in parallel, with all laboratories successfully detecting the 0.5% JAK2-V617F dilution (with Ct values <40 cycles recorded by all laboratories with each assay). The Ct values observed with neat K562 cells (labeled as 0% JAK2-V617F) generally exceeded 40 cycles (with the exception of laboratory 2 and laboratory 4 for Assay 2 and Assay 7, respectively), consistent with specificity of the JAK2-V617F assays for the mutant allele with the test conditions applied in the majority of the laboratories.
Following a further QC round, Cyclophilin was dropped, as reproducibility was found to be poorer as compared with the ALB assay, which is widely used as a control gene in the MRD detection of acute lymphoblastic leukemia. With the aim of selecting the best assays for the tracking of MRD (%JAK2-V617F <1), in QC5, two further ‘in-house’ assays were evaluated (Assays 8 and 9), but were eliminated because they exhibited poorer specificity when tested on serial dilutions of HEL in K562 cells. On the basis of two more QC rounds, Assays 2 and 6 were also eliminated; the former generally afforded poorer specificity, whereas the latter was less efficient (slope—4.21, y intercept 51.5), rendering them less suitable for MRD monitoring (Figure 2) as compared with Assays 5 and 7 (slopes—3.5, y intercept 41, Figure 2) for which the majority of laboratories detected the 0.08% dilution (Table 1).
Evaluation of specificity of JAK2-V617F assays in control PB samples
Although the two best-performing JAK2-V617F assays showed limited cross-reactivity with the wild-type allele on analysis of undiluted K562 cells, with most laboratories reporting Ct values >40 (see plots for 0% JAK2-V617F for Assays 5 and 7, Figure 3), we subsequently formally evaluated assay specificity in a large number of PB samples (n=100) derived from individuals with no history of MPN. For these experiments, ALB was used to provide an independent reference to control for the amount of DNA template, with the level of background amplification with the JAK2-V617F assay assessed by the ΔCt method. Some cross-reactivity was observed with Assay 7, with Ct values of <40 observed in the majority (54%), with median Ct 39.9 (range 38.6–50). Assay 5 exhibited less cross-reactivity, with Ct values <40 cycles observed in only 3% of samples (median 40.9, range 39.9–50.0) (Supplementary Figure 3). Accordingly, there was a greater difference in ΔCtJAK2-V617F-ALB for Assay 5 (median 14.1, range 13.3–21.6) as compared with Assay 7 (median 13.2, range 11.7–21.6), suggesting that Assay 5 may have a marginally better specificity and performance profile for MRD assessment, more reliably distinguishing low level residual disease from non-specific background.
The two best-performing JAK2-V617F assays for MRD tracking predict outcome following allogeneic transplant
To assess the reliability of the most sensitive assays (Assays 5 and 7) to detect MRD and to predict outcome, they were evaluated in stored samples from a cohort of 28 patients with a range of JAK2-V617F-positive myeloid neoplasms (including PMF, PV, transformed MPN and chronic myelomonocytic leukemia) who underwent allogeneic stem cell transplantation, mostly with reduced intensity conditioning given the age of the patients concerned (median 54 years). Overall, comparison of the results obtained with the two assays in pre- and post-transplant samples showed very close concordance—r=0.98, P<0.0001 (Supplementary Figure 4). All cases tested positive for JAK2-V617F immediately before transplant, with 17 patients achieving molecular remission at a median of 108 days (range 23–1488 days). Five patients remained in molecular remission at date of last follow-up (3–25 months post-transplant), testing negative for JAK2-V617F without any fluctuation or background amplification. In 12 patients after initial molecular remission, the qPCR assays detected recurrence of JAK2-V617F positivity; this predicted clinical relapse in 9 patients 1.5–19.6 months later, predating the loss of full donor chimerism by 1.5–12.2 months (Figure 4). Three patients with PCR positivity were still reported to be in clinical remission at the time of last follow-up (3–8 months later). Clinical relapses were predicted by prior JAK2-V617F positivity by the qPCR assays, with the exception of one case, where there was a prolonged interval (10 months) between relapse and the preceding MRD assessment. The kinetics of relapse was highly variable, with the JAK2-V617F mutant allele rising by a median of 0.6 logs/month (range 0.1–1.3 logs/month). In seven patients, donor lymphocyte infusion (DLI) was administered for clinical relapse (myelofibrosis, n=4, transformed MPN, n=3) leading to disease response in four patients (three myelofibrosis, one transformed MPN), associated with the achievement of molecular remission by qPCR (Figure 4).
Investigation of best-performing JAK2-V617F qPCR assays to predict outcome following allogeneic transplantation. Stored PB samples from patients with JAK2-V617F-positive myeloid disorders were tested in parallel using JAK2-V617F Assays 5 and 7, in conjunction with ALB as an independent control gene. Samples were determined to be positive or negative for JAK2-V617F according to the ΔCt between the JAK2-V617F and ALB amplification plots, taking into account the cut offs defined in control PB samples for each mutant assay (see Supplementary Figure 3), with PCR-positive and -negative samples denoted by filled and empty data points, respectively. JAK2-V617F percentage for PCR-positive samples was calculated using the matching wild-type and total JAK2 assays for the respective assays. PCR profiles are shown from four (UPN1–4) of the 28 patients evaluated, who underwent allogeneic transplant with reduced intensity conditioning with an unrelated (UPN1) or sibling donor (UPN2–4) for PMF (UPN2, UPN3) or JAK2-V617F-positive secondary acute myeloid leukemia arising on a background of MPN (UPN1, UPN4). In each case, the clinical relapse post-transplant was predicted by JAK2-V617F positivity with a rise in mutant level (UPN1, UPN3 and UPN4) or persistently high levels (UPN2). JAK2-V617F detection by qPCR preceded loss of donor chimerism in UPN1, UPN3 and UPN4. Three patients received DLI, denoted by blue arrows, inducing molecular remission. Chimerism was assessed by microsatellite marker analysis in UPN1, UPN3 and UPN4 and by fluorescence in situ hybridization analysis of X and Y chromosomes in UPN2.
Discussion
The advent of high throughput sequencing technologies has led to major advances in defining the mutational landscape of hematological malignancies. This information has not only helped refine disease diagnosis and discern potential therapeutic targets, but also provides opportunities to develop assays to detect MRD, tracking response to therapy. Extensive standardization efforts have been undertaken to establish optimal qPCR assays to detect leukemia-specific targets, for example, BCR-ABL1 and other fusion gene transcripts arising from balanced chromosomal rearrangements, as well as immunoglobulin and T-cell receptor gene rearrangements in lymphoid malignancies.46, 48 These initiatives have been highly influential, leading to the implementation of standardized assays, which have been shown to provide independent prognostic information, to guide therapy within clinical trials. However, a number of hematological malignancies are characterized by point mutations, for example, BRAF-V600E in hairy cell leukemia and JAK2-V617F in MPNs, which can also potentially be detected and tracked by sensitive qPCR assays to predict treatment outcome.26, 27, 28, 29, 49 Point mutations present more challenging MRD targets as compared with leukemic fusion genes due to potential problems relating to cross-reactivity of the mutant assay with the wild-type allele (reviewed12), with a previous study showing that the most sensitive and accurate approach for detection of JAK2-V617F involves qPCR with mutation-specific primers.10
For patients with suspected MPNs, there is a critical need for robust and sensitive assays to detect JAK2-V617F, with mutant allele burden reported to correlate with biologically and prognostically distinct subsets of disease (reviewed6). Importantly, it has become clear that the level of JAK2-V617F detectable in the PB of some patients with bona fide MPNs can be as low as 1% at the time of diagnosis,6, 7, 8, 9, 10 which is below the detection limit of some standard diagnostic methods, such as pyrosequencing.10, 11 Since the discovery of the JAK2-V617F mutation, numerous qPCR assays have been designed to detect this abnormality,10, 12, 50 which we have shown vary markedly in their sensitivity and specificity. Previous studies have highlighted the potential of qPCR assessment of JAK2-V617F allele burden to predict outcome following allogeneic transplant in PMF,26, 27, 28, 29 with reduction of JAK2-V617F to <1% by 1 month30 or achievement of PCR negativity in the PB by 6 months post-transplant, distinguishing patients at markedly differing risk of subsequent disease relapse.29 However, these time points were not particularly informative in the present study, which may reflect the greater heterogeneity of the patients analyzed.
Previous reports have highlighted the importance of serial MRD assessment post-transplant as a potential tool to guide immunosuppression and administration of DLI.29, 30, 31, 32 We observed that DLI given at time of clinical relapse induced molecular remission as determined by qPCR in four patients (Figure 4), lending further support for a graft vs MPN effect. Chimerism analysis currently provides the mainstay for post-transplant surveillance, with loss of donor chimerism used to direct DLI and reduce the risk of disease relapse. However, chimerism assays do not directly assess the presence of MRD and are relatively insensitive. Indeed, in the present study we found that qPCR assays detected JAK2-V617F positivity long before loss of full donor chimerism and clinical relapse. In accordance with the experience in chronic myeloid leukemia, where DLI has been shown to be more effective in the context of MRD as compared with cytogenetic or hematological relapse (reviewed51), there is evidence to suggest that early intervention with DLI in the context of molecular relapse of JAK2-V617F-associated PMF post-transplant is associated with a better response rate compared with salvage at time of overt relapse.31 Therefore, there is now a strong rationale for the adoption of regular qPCR assessment for JAK2-V617F following allogeneic transplant in informative patients to complement chimerism analysis.52
Although results obtained with Assays 5 and 7 in pre- and post-transplant patient samples were found to be highly concordant (Supplementary Figure 4), the assessment of control PB samples suggested that Assay 5, previously published by Larsen et al.43 may more reliably distinguish low level MRD from background amplification. Indeed, this assay has been applied in the diagnostic setting in a large population-based study to discriminate JAK2-V617F status,53 following a Chinese study that reported the detection of low level JAK2-V617F positivity in individuals with no clinical evidence of an MPN.54 Here, we significantly extend this study, showing that the selected ELN/MPN&MPNr-EuroNet JAK2-V617F assay (Assay 5) can be used to reliably track MRD, suitable for guiding management post-transplant. Importantly, we found that this assay performed well across different qPCR platforms (Table 1), giving rise to comparable results, and therefore can be recommended for assessing the impact of novel therapies on the malignant clone in clinical trials, as well being implemented into the routine management of patients undergoing allogeneic transplantation. The recommended qPCR assay should provide a benchmark against which to assess sensitivity and specificity of high-throughput sequencing approaches to detect point mutations such as JAK2-V617F, which promise to extend the molecular detection of MRD to much greater proportions of patients with hematological malignancies, thereby allowing development of more personalized treatment approaches.
References
James C, Ugo V, Le Couédic JP, Staerk J, Delhommeau F, Lacout C et al. A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature 2005; 434: 1144–1148.
Kralovics R, Passamonti F, Buser AS, Teo SS, Tiedt R, Passweg JR et al. A gain-of-function mutation of JAK2 in myeloproliferative disorders. N Engl J Med 2005; 352: 1779–1790.
Levine RL, Wadleigh M, Cools J, Ebert BL, Wernig G, Huntly BJ et al. Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell 2005; 7: 387–397.
Baxter EJ, Scott LM, Campbell PJ, East C, Fourouclas N, Swanton S et al. Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet 2005; 365: 1054–1061.
Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H et al. (eds). WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. IARC: Lyon, France, 2008.
Cross NC . Genetic and epigenetic complexity in myeloproliferative neoplasms. Hematology Am Soc Hematol Educ Program 2011, 208–214.
Kittur J, Knudson RA, Lasho TL, Finke CM, Gangat N, Wolanskyj AP et al. Clinical correlates of JAK2V617F allele burden in essential thrombocythemia. Cancer 2007; 109: 2279–2284.
Wang YL, Vandris K, Jones A, Cross NC, Christos P, Adriano F et al. JAK2 mutations are present in all cases of polycythemia vera. Leukemia 2008; 22: 1289.
Mason J, Akiki S, Griffiths MJ . Pitfalls in molecular diagnosis in haemato-oncology. J Clin Pathol 2011; 64: 275–278.
Lippert E, Girodon F, Hammond E, Jelinek J, Reading NS, Fehse B et al. Concordance of assays designed for the quantification of JAK2V617F: a multicenter study. Haematologica 2009; 94: 38–45.
Bench AJ, White HE, Foroni L, Godfrey AL, Gerrard G, Akiki S et al. Molecular diagnosis of the myeloproliferative neoplasms: UK guidelines for the detection of JAK2 V617F and other relevant mutations. Br J Haematol 2013; 160: 25–34.
Bench AJ . The role of molecular genetic analysis within the diagnostic haemato-oncology laboratory. Int J Lab Hematol 2012; 34: 21–34.
Tefferi A, Strand JJ, Lasho TL, Elliott MA, Li CY, Mesa RA et al. Respective clustering of unfavorable and favorable cytogenetic clones in myelofibrosis with myeloid metaplasia with homozygosity for JAK2(V617F) and response to erythropoietin therapy. Cancer 2006; 106: 1739–1743.
Vannucchi AM, Antonioli E, Guglielmelli P, Rambaldi A, Barosi G, Marchioli R et al. Clinical profile of homozygous JAK2 617V>F mutation in patients with polycythemia vera or essential thrombocythemia. Blood 2007; 110: 840–846.
Barosi G, Bergamaschi G, Marchetti M, Vannucchi AM, Guglielmelli P, Antonioli E et al. JAK2 V617F mutational status predicts progression to large splenomegaly and leukemic transformation in primary myelofibrosis. Blood 2007; 110: 4030–4036.
Vannucchi AM, Antonioli E, Guglielmelli P, Longo G, Pancrazzi A, Ponziani V et al. Prospective identification of high-risk polycythemia vera patients based on JAK2(V617F) allele burden. Leukemia 2007; 21: 1952–1959.
Tefferi A, Lasho TL, Huang J, Finke C, Mesa RA, Li CY et al. Low JAK2V617F allele burden in primary myelofibrosis, compared to either a higher allele burden or unmutated status, is associated with inferior overall and leukemia-free survival. Leukemia 2008; 22: 756–761.
Guglielmelli P, Barosi G, Specchia G, Rambaldi A, Lo Coco F, Antonioli E et al. Identification of patients with poorer survival in primary myelofibrosis based on the burden of JAK2V617F mutated allele. Blood 2009; 114: 1477–1483.
De Stefano V, Za T, Rossi E, Fiorini A, Ciminello A, Luzzi C et al. Influence of the JAK2 V617F mutation and inherited thrombophilia on the thrombotic risk among patients with essential thrombocythemia. Haematologica 2009; 94: 733–737.
Passamonti F, Rumi E, Pietra D, Elena C, Boveri E, Arcaini L et al. A prospective study of 338 patients with polycythemia vera: the impact of JAK2 (V617F) allele burden and leukocytosis on fibrotic or leukemic disease transformation and vascular complications. Leukemia 2010; 24: 1574–1579.
Kiladjian JJ, Cassinat B, Turlure P, Cambier N, Roussel M, Bellucci S et al. High molecular response rate of polycythemia vera patients treated with pegylated interferon alpha-2a. Blood 2006; 108: 2037–2040.
Jabbour E, Kantarjian H, Cortes J, Thomas D, Garcia-Manero G, Ferrajoli A et al. PEG-IFN-alpha-2b therapy in BCR-ABL-negative myeloproliferative disorders: final result of a phase 2 study. Cancer 2007; 110: 2012–2018.
Kiladjian JJ, Cassinat B, Chevret S, Turlure P, Cambier N, Roussel M et al. Pegylated interferon-alfa-2a induces complete hematologic and molecular responses with low toxicity in polycythemia vera. Blood 2008; 112: 3065–3072.
Quintás-Cardama A, Kantarjian H, Manshouri T, Luthra R, Estrov Z, Pierce S et al. Pegylated interferon alfa-2a yields high rates of hematologic and molecular response in patients with advanced essential thrombocythemia and polycythemia vera. J Clin Oncol 2009; 27: 5418–5424.
Larsen TS, Bjerrum OW, Pallisgaard N, Andersen MT, Møller MB, Hasselbalch HC . Sustained major molecular response on interferon alpha-2b in two patients with polycythemia vera. Ann Hematol 2008; 87: 847–850.
Kröger N, Badbaran A, Holler E, Hahn J, Kobbe G, Bornhäuser M et al. Monitoring of the JAK2-V617F mutation by highly sensitive quantitative real-time PCR after allogeneic stem cell transplantation in patients with myelofibrosis. Blood 2007; 109: 1316–1321.
Steckel NK, Koldehoff M, Ditschkowski M, Beelen DW, Elmaagacli AH . Use of the activating gene mutation of the tyrosine kinase (VAL617Phe) JAK2 as a minimal residual disease marker in patients with myelofibrosis and myeloid metaplasia after allogeneic stem cell transplantation. Transplantation 2007; 83: 1518–1520.
Siebolts U, Lange T, Niederwieser D, Wickenhauser C . Allele-specific wild-type blocker quantitative PCR for highly sensitive detection of rare JAK2 p.V617F point mutation in primary myelofibrosis as an appropriate tool for the monitoring of molecular remission following therapy. J Clin Path 2010; 63: 370–372.
Alchalby H, Badbaran A, Zabelina T, Kobbe G, Hahn J, Wolff D et al. Impact of JAK2V617F mutation status, allele burden, and clearance after allogeneic stem cell transplantation for myelofibrosis. Blood 2010; 116: 3572–3581.
Lange T, Edelmann A, Siebolts U, Krahl R, Nehring C, Jäkel N et al. JAK2 p.V617F allele burden in myeloproliferative neoplasms one month after allogeneic stem cell transplantation significantly predicts outcome and risk of relapse. Haematologica 2013; 98: 722–728.
Kröger N, Alchalby H, Klyuchnikov E, Badbaran A, Hildebrandt Y, Ayuk F et al. JAK2-V617F-triggered preemptive and salvage adoptive immunotherapy with donor-lymphocyte infusion in patients with myelofibrosis after allogeneic stem cell transplantation. Blood 2009; 113: 1866–1868.
Benjamini O, Koren-Michowitz M, Amariglio N, Kröger N, Nagler A, Shimoni A . Relapse of postpolycythemia myelofibrosis after allogeneic stem cell transplantation in a polycythemic phase: successful treatment with donor lymphocyte infusion directed by quantitative PCR test for V617F-JAK2 mutation. Leukemia 2008; 22: 1961–1963.
Verstovsek S, Kantarjian H, Mesa RA, Pardanani AD, Cortes-Franco J, Thomas DA et al. Safety and efficacy of INCB018424, a JAK1 and JAK2 inhibitor, in myelofibrosis. N Engl J Med 2010; 363: 1117–1127.
Santos FP, Kantarjian HM, Jain N, Manshouri T, Thomas DA, Garcia-Manero G et al. Phase 2 study of CEP-701, an orally available JAK2 inhibitor, in patients with primary or post-polycythemia vera/essential thrombocythemia myelofibrosis. Blood 2010; 115: 1131–1136.
Pardanani A, Gotlib JR, Jamieson C, Cortes JE, Talpaz M, Stone RM et al. Safety and efficacy of TG101348, a selective JAK2 inhibitor, in myelofibrosis. J Clin Oncol 2011; 29: 789–796.
Verstovsek S, Mesa RA, Gotlib J, Levy RS, Gupta V, DiPersio JF et al. A double-blind, placebo-controlled trial of ruxolitinib for myelofibrosis. N Engl J Med 2012; 366: 799–807.
Baerlocher GM, Oppliger Leibundgut E, Ayran C, Blaney M, Burington B, Morfeld D et al. Imetelstat rapidly induces and maintains substantial hematologic and molecular responses in patients with essential thrombocythemia (ET) who are refractory or intolerant to prior therapy: Preliminary phase II results. Congr Am Soc Hematol 2012; Abstract 179.
Spindler KL, Pallisgaard N, Vogelius I, Jakobsen A . Quantitative cell-free DNA, KRAS, and BRAF mutations in plasma from patients with metastatic colorectal cancer during treatment with cetuximab and irinotecan. Clin Cancer Res 2012; 18: 1177–1185.
Pongers-Willemse MJ, Verhagen OJHM, Tibbe GJM, Wijkhuijs AJM, de Haas V, Roovers E et al. Real-time quantitative PCR for the detection of minimal residual disease in acute lymphoblastic leukemia using junctional region specific TaqMan probes. Leukemia 1998; 12: 2006–2014.
Levine RL, Belisle C, Wadleigh M, Zahrieh D, Lee S, Chagnon P et al. X-inactivation-based clonality analysis and quantitative JAK2V617F assessment reveal a strong association between clonality and JAK2V617F in PV but not ET/MMM, and identifies a subset of JAK2V617F-negative ET and MMM patients with clonal hematopoiesis. Blood 2006; 107: 4139–4141.
Lippert E, Boissinot M, Kralovics R, Girodon F, Dobo I, Praloran V et al. The JAK2-V617F mutation is frequently present at diagnosis in patients with essential thrombocythemia and polycythemia vera. Blood 2006; 108: 1865–1867.
Ishii T, Bruno E, Hoffman R, Xu M . Involvement of various hematopoietic-cell lineages by the JAK2V617F mutation in polycythemia vera. Blood 2006; 108: 3128–3134.
Larsen TS, Christensen JH, Hasselbalch HC, Pallisgaard N . The JAK2 V617F mutation involves B- and T-lymphocyte lineages in a subgroup of patients with Philadelphia-chromosome negative chronic myeloproliferative disorders. Br J Haematol 2007; 136: 745–751.
Nussenzveig RH, Swierczek SI, Jelinek J, Gaikwad A, Liu E, Verstovsek S et al. Polycythemia vera is not initiated by JAK2V617F mutation. Exp Hematol 2007; 35: 32–38.
Flora R, Grimwade D . Real-time quantitative RT-PCR to detect fusion gene transcripts associated with AML. Methods Mol Med 2004; 91: 151–173.
Gabert J, Beillard E, van der Velden VH, Bi W, Grimwade D, Pallisgaard N et al. Standardization and quality control studies of 'real-time' quantitative reverse transcriptase polymerase chain reaction of fusion gene transcripts for residual disease detection in leukemia - a Europe Against Cancer program. Leukemia 2003; 17: 2318–2357.
Grimwade D, Jovanovic JV, Hills RK, Nugent EA, Patel Y, Flora R et al. Prospective minimal residual disease monitoring to predict relapse of acute promyelocytic leukemia and to direct pre-emptive arsenic trioxide therapy. J Clin Oncol 2009; 27: 3650–3658.
van der Velden VH, Panzer-Grümayer ER, Cazzaniga G, Flohr T, Sutton R, Schrauder A et al. Optimization of PCR-based minimal residual disease diagnostics for childhood acute lymphoblastic leukemia in a multi-center setting. Leukemia 2007; 21: 706–713.
Schnittger S, Bacher U, Haferlach T, Wendland N, Ulke M, Dicker F et al. Development and validation of a real-time quantification assay to detect and monitor BRAFV600E mutations in hairy cell leukemia. Blood 2012; 119: 3151–3154.
Merker JD, Jones CD, Oh ST, Schrijver I, Gotlib J, Zehnder JL . Design and evaluation of a real-time PCR assay for quantification of JAK2 V617F and wild-type JAK2 transcript levels in the clinical laboratory. J Mol Diagn 2010; 12: 58–64.
Dazzi F, Szydlo RM, Goldman JM . Donor lymphocyte infusions for relapse of chronic myeloid leukemia after allogeneic stem cell transplant: where we now stand. Exp Hematol 1999; 27: 1477–1486.
Reilly JT, McMullin MF, Beer PA, Butt N, Conneally E, Duncombe A et al. Guideline for the diagnosis and management of myelofibrosis. Br J Haematol 2012; 158: 453–471.
Nielsen C, Birgens HS, Nordestgaard BG, Bojesen SE . Diagnostic value of JAK2 V617F somatic mutation for myeloproliferative cancer in 49 488 individuals from the general population. Br J Haematol 2013; 160: 70–79.
Xu X, Zhang Q, Luo J, Xing S, Li Q, Krantz SB et al. JAK2(V617F): Prevalence in a large Chinese hospital population. Blood 2007; 109: 339–342.
Acknowledgements
This work was supported by the European LeukemiaNet; DG, BW, KT and CNH gratefully acknowledge research funding from the Guy’s and St Thomas’ Charity. The EU Co-operation in Science and Technology (COST) program supports MPN&MPNr-EuroNet (COST Action BM0902). We wish to thank Marina Migeon for expert technical assistance. We are grateful to Central England Haemato-Oncology Research Biobank for provision of primary patient samples. We are also grateful to Prof John Reilly and UK NEQAS for providing freeze-dried cell line dilutions for QC analysis within the ELN.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Nicolas Maroc is an employee of Ipsogen (now Qiagen Marseille); however, Ipsogen were not involved in the data analyses which led to the selection of the optimal JAK2-V617F qPCR assay reported in this study. The remaining authors declare no conflict of interest.
Additional information
These data were presented in part at the 53rd American Society of Hematology meeting, San Diego, December 2011.
Supplementary Information accompanies this paper on the Leukemia website
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Jovanovic, J., Ivey, A., Vannucchi, A. et al. Establishing optimal quantitative-polymerase chain reaction assays for routine diagnosis and tracking of minimal residual disease in JAK2-V617F-associated myeloproliferative neoplasms: a joint European LeukemiaNet/MPN&MPNr-EuroNet (COST action BM0902) study. Leukemia 27, 2032–2039 (2013). https://doi.org/10.1038/leu.2013.219
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2013.219
Keywords
This article is cited by
-
JAK2V617F variant allele frequency, non-driver mutations, single-nucleotide variants and polycythemia vera outcome
Journal of Cancer Research and Clinical Oncology (2023)
-
Clonogenic assays improve determination of variant allele frequency of driver mutations in myeloproliferative neoplasms
Annals of Hematology (2022)
-
Essential thrombocythaemia progression to the fibrotic phase is associated with a decrease in JAK2 and PDL1 levels
Annals of Hematology (2022)
-
Clonal hematopoiesis with JAK2V617F promotes pulmonary hypertension with ALK1 upregulation in lung neutrophils
Nature Communications (2021)
-
Rapid, low cost and sensitive detection of Calreticulin mutations by a PCR based amplicon length differentiation assay for diagnosis of myeloproliferative neoplasms
BMC Medical Genetics (2019)