Abstract
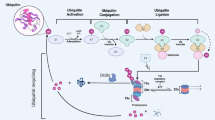
The delicate balance between the synthesis and the degradation of proteins ensures cellular homeostasis. Proteases act in an irreversible manner and therefore have to be strictly regulated. The ubiquitin–proteasome system (UPS) is a major pathway for the proteolytic degradation of cellular proteins. As dysregulation of the UPS is observed in most cancers including leukemia, the UPS is a valid target for therapeutic intervention strategies. Ubiquitin-ligases selectively bind substrates to target them for poly-ubiquitinylation and proteasomal degradation. Therefore, pharmacological modulation of these proteins could allow a specific level of control. Increasing evidence accumulates that ubiquitin-ligases termed mammalian seven in absentia homologs (SIAHs) are not only critical for the pathogenesis of solid tumors but also for leukemogenesis. However, the relevance and therapeutic potential of SIAH-dependent processes has not been fully elucidated. Here, we summarize functions of SIAH ubiquitin-ligases in leukemias, how they select leukemia-relevant substrates for proteasomal degradation, and how the expression and activity of SIAH1 and SIAH2 can be modulated in vivo. We also discuss that epigenetic drugs belonging to the group of histone deacetylase inhibitors induce SIAH-dependent proteasomal degradation to accelerate the turnover of leukemogenic proteins. In addition, our review highlights potential areas for future research on SIAH proteins.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Buchwald M, Krämer OH, Heinzel T . HDACi–targets beyond chromatin. Cancer Lett 2009; 280: 160–167.
Hoeller D, Dikic I . Targeting the ubiquitin system in cancer therapy. Nature 2009; 458: 438–444.
Bedford L, Lowe J, Dick LR, Mayer RJ, Brownell JE . Ubiquitin-like protein conjugation and the ubiquitin-proteasome system as drug targets. Nat Rev Drug Discov 2011; 10: 29–46.
Wenzel DM, Stoll KE, Klevit RE . E2s: structurally economical and functionally replete. Biochem J 2011; 433: 31–42.
Martens JH, Stunnenberg HG . The molecular signature of oncofusion proteins in acute myeloid leukemia. FEBS Lett 2010; 584: 2662–2669.
Shah JJ, Orlowski RZ . Proteasome inhibitors in the treatment of multiple myeloma. Leukemia 2009; 23: 1964–1979.
Mohty B, El-Cheikh J, Yakoub-Agha I, Avet-Loiseau H, Moreau P, Mohty M . Treatment strategies in relapsed and refractory multiple myeloma: a focus on drug sequencing and ‘retreatment’ approaches in the era of novel agents. Leukemia 2012; 26: 73–85.
Müller S, Krämer OH . Inhibitors of HDACs—effective drugs against cancer? Curr Cancer Drug Targets 2010; 10: 210–228.
Quintás-Cardama A, Santos FP, Garcia-Manero G . Histone deacetylase inhibitors for the treatment of myelodysplastic syndrome and acute myeloid leukemia. Leukemia 2011; 25: 226–235.
House CM, Möller A, Bowtell DD . Siah proteins: novel drug targets in the Ras and hypoxia pathways. Cancer Res 2009; 69: 8835–8838.
Qi J, Pellecchia M, Ronai ZA . The Siah2-HIF-FoxA2 axis in prostate cancer—new markers and therapeutic opportunities. Oncotarget 2010; 1: 379–385.
Bursen A, Moritz S, Gaussmann A, Dingermann T, Marschalek R . Interaction of AF4 wild-type and AF4.MLL fusion protein with SIAH proteins: indication for t4;11 pathobiology? Oncogene 2004; 23: 6237–6249.
Liu M, Hsu J, Chan C, Li Z, Zhou Q . The ubiquitin ligase Siah1 controls ELL2 stability and formation of super elongation complexes to modulate gene transcription. Mol Cell 2012; 46: 325–334.
Mei Y, Xie C, Xie W, Wu Z, Wu M . Siah-1S, a novel splice variant of Siah-1 seven in absentia homolog, counteracts Siah-1-mediated downregulation of beta-catenin. Oncogene 2007; 26: 6319–6331.
Buchwald M, Pietschmann K, Müller JP, Böhmer FD, Heinzel T, Krämer OH . Ubiquitin conjugase UBCH8 targets active FMS-like tyrosine kinase 3 for proteasomal degradation. Leukemia 2010; 24: 1412–1421.
Pietschmann K, Buchwald M, Müller S, Knauer SK, Kögl M, Heinzel T et al. Differential regulation of PML-RARalpha stability by the ubiquitin ligases SIAH1/SIAH2 and TRIAD1. Int J Biochem Cell Biol 2012; 44: 132–138.
Santelli E, Leone M, Li C, Fukushima T, Preece NE, Olson AJ et al. Structural analysis of Siah1-Siah-interacting protein interactions and insights into the assembly of an E3 ligase multiprotein complex. J Biol Chem 2005; 280: 34278–34287.
Fukushima T, Zapata JM, Singha NC, Thomas M, Kress CL, Krajewska M et al. Critical function for SIP, a ubiquitin E3 ligase component of the beta-catenin degradation pathway, for thymocyte development and G1 checkpoint. Immunity 2006; 24: 29–39.
Nagano Y, Fukushima T, Okemoto K, Tanaka K, Bowtell DD, Ronai Z et al. Siah1/SIP regulates p27kip1 stability and cell migration under metabolic stress. Cell Cycle 2011; 10: 2592–2602.
Xu Z, Sproul A, Wang W, Kukekov N, Greene LA . Siah1 interacts with the scaffold protein POSH to promote JNK activation and apoptosis. J Biol Chem 2006; 281: 303–312.
Wu H, Shi Y, Lin Y, Qian W, Yu Y, Huo K . Eukaryotic translation elongation factor 1 delta inhibits the ubiquitin ligase activity of SIAH-1. Mol Cell Biochem 2011; 357: 209–215.
Wu H, Lin Y, Shi Y, Qian W, Tian Z, Yu Y et al. SIAH-1 interacts with mammalian polyhomeotic homologues HPH2 and affects its stability via the ubiquitin-proteasome pathway. Biochem Biophys Res Commun 2010; 397: 391–396.
Telerman A, Amson R . The molecular programme of tumour reversion: the steps beyond malignant transformation. Nat Rev Cancer 2009; 9: 206–216.
Chan P, Möller A, Liu MC, Sceneay JE, Wong CS, Waddell N et al. The expression of the ubiquitin ligase SIAH2 seven in absentia homolog 2 is mediated through gene copy number in breast cancer and is associated with a basal-like phenotype and p53 expression. Breast Cancer Res 2011; 13: R19.
Frew IJ, Dickins RA, Cuddihy AR, Del Rosario M, Reinhard C, O'Connell MJ et al. Normal p53 function in primary cells deficient for Siah genes. Mol Cell Biol 2002; 22: 8155–8164.
Xie W, Jin L, Mei Y, Wu M . E2F1 represses beta-catenin/TCF activity by direct up-regulation of Siah1. J Cell Mol Med 2009; 13: 1719–1727.
Frasor J, Danes JM, Funk CC, Katzenellenbogen BS . Estrogen down-regulation of the corepressor N-CoR: mechanism and implications for estrogen derepression of N-CoR-regulated genes. Proc Natl Acad Sci USA 2005; 102: 13153–13157.
Depaux A, Regnier-Ricard F, Germani A, Varin-Blank N . Dimerization of hSiah proteins regulates their stability. Biochem Biophys Res Commun 2006; 348: 857–863.
Scortegagna M, Subtil T, Qi J, Kim H, Zhao W, Gu W et al. USP13 enzyme regulates Siah2 ligase stability and activity via noncatalytic ubiquitin-binding domains. J Biol Chem 2011; 286: 27333–27341.
Shah M, Stebbins JL, Dewing A, Qi J, Pellecchia M, Ronai ZA . Inhibition of Siah2 ubiquitin ligase by vitamin K3 menadione attenuates hypoxia and MAPK signaling and blocks melanoma tumorigenesis. Pigment Cell Melanoma Res 2009; 22: 799–808.
Calzado MA, de la Vega L, Möller A, Bowtell DD, Schmitz ML . An inducible autoregulatory loop between HIPK2 and Siah2 at the apex of the hypoxic response. Nat Cell Biol 2009; 11: 85–91.
Winter M, Sombroek D, Dauth I, Moehlenbrink J, Scheuermann K, Crone J et al. Control of HIPK2 stability by ubiquitin ligase Siah-1 and checkpoint kinases ATM and ATR. Nat Cell Biol 2008; 10: 812–824.
Kim SY, Choi DW, Kim EA, Choi CY . Stabilization of HIPK2 by escape from proteasomal degradation mediated by the E3 ubiquitin ligase Siah1. Cancer Lett 2009; 279: 177–184.
Khurana A, Nakayama K, Williams S, Davis RJ, Mustelin T, Ronai Z . Regulation of the ring finger E3 ligase Siah2 by p38 MAPK. J Biol Chem 2006; 281: 35316–35326.
Krämer OH, Müller S, Buchwald M, Reichardt S, Heinzel T . Mechanism for ubiquitylation of the leukemia fusion proteins AML1-ETO and PML-RARalpha. Faseb J 2008; 22: 1369–1379.
Krämer OH . HDAC2: a critical factor in health and disease. Trends Pharmacol Sci 2009; 30: 647–655.
Frew IJ, Hammond VE, Dickins RA, Quinn JM, Walkley CR, Sims NA et al. Generation and analysis of Siah2 mutant mice. Mol Cell Biol 2003; 23: 9150–9161.
Lipkowitz S, Weissman AM . RINGs of good and evil: RING finger ubiquitin ligases at the crossroads of tumour suppression and oncogenesis. Nat Rev Cancer 2011; 11: 629–643.
Liu M, Aneja R, Wang H, Sun L, Dong X, Huo L et al. Modulation of multidrug resistance in cancer cells by the E3 ubiquitin ligase seven-in-absentia homologue 1. J Pathol 2008; 214: 508–514.
Ahmed AU, Schmidt RL, Park CH, Reed NR, Hesse SE, Thomas CF et al. Effect of disrupting seven-in-absentia homolog 2 function on lung cancer cell growth. J Natl Cancer Inst 2008; 100: 1606–1629.
Matsuo K, Satoh S, Okabe H, Nomura A, Maeda T, Yamaoka Y et al. SIAH1 inactivation correlates with tumor progression in hepatocellular carcinomas. Genes Chromosomes Cancer 2003; 36: 283–291.
Kim CJ, Cho YG, Park CH, Jeong SW, Nam SW, Kim SY et al. Inactivating mutations of the Siah-1 gene in gastric cancer. Oncogene 2004; 23: 8591–8596.
Marschalek R . Mechanisms of leukemogenesis by MLL fusion proteins. Br J Haematol 2011; 152: 141–154.
Pless B, Oehm C, Knauer S, Stauber RH, Dingermann T, Marschalek R . The heterodimerization domains of MLL-FYRN and FYRC--are potential target structures in t4;11 leukemia. Leukemia 2011; 25: 663–670.
Benedikt A, Baltruschat S, Scholz B, Bursen A, Arrey TN, Meyer B et al. The leukemogenic AF4-MLL fusion protein causes P-TEFb kinase activation and altered epigenetic signatures. Leukemia 2011; 25: 135–144.
Smith E, Lin C, Shilatifard A . The super elongation complex SEC and MLL in development and disease. Genes Dev 2011; 25: 661–672.
Montes R, Ayllón V, Gutierrez-Aranda I, Prat I, Hernández-Lamas MC, Ponce L et al. Enforced expression of MLL-AF4 fusion in cord blood CD34+ cells enhances the hematopoietic repopulating cell function and clonogenic potential but is not sufficient to initiate leukemia. Blood 2011; 117: 4746–4758.
Hsieh JJ, Cheng EH, Korsmeyer SJ . Taspase1: a threonine aspartase required for cleavage of MLL and proper HOX gene expression. Cell 2003; 115: 293–303.
Meyer C, Kowarz E, Hofmann J, Renneville A, Zuna J, Trka J et al. New insights to the MLL recombinome of acute leukemias. Leukemia 2009; 23: 1490–1499.
Tan J, Muntean AG, Hess JL . PAFc, a key player in MLL-rearranged leukemogenesis. Oncotarget 2010; 1: 461–465.
Balgobind BV, Zwaan CM, Pieters R, Van den Heuvel-Eibrink MM . The heterogeneity of pediatric MLL-rearranged acute myeloid leukemia. Leukemia 2011; 25: 1239–1248.
Bursen A, Schwabe K, Ruster B, Henschler R, Ruthardt M, Dingermann T et al. The AF4.MLL fusion protein is capable of inducing ALL in mice without requirement of MLL.AF4. Blood 2010; 115: 3570–3579.
Takeda S, Chen DY, Westergard TD, Fisher JK, Rubens JA, Sasagawa S et al. Proteolysis of MLL family proteins is essential for taspase1-orchestrated cell cycle progression. Genes Dev 2006; 20: 2397–2409.
Bier C, Knauer SK, Klapthor A, Schweitzer A, Rekik A, Krämer OH et al. Cell-based analysis of structure-function activity of threonine aspartase 1. J Biol Chem 2011; 286: 3007–3017.
Bier C, Knauer SK, Wünsch D, Kunst L, Scheiding S, Kaiser M et al. Allosteric inhibition of Taspase1’s pathobiological activity by enforced dimerization in vivo. Faseb J 2012; 26: 3421–3429.
Bier C, Knauer SK, Docter D, Schneider G, Krämer OH, Stauber RH . The importin-alpha/nucleophosmin switch controls taspase1 protease function. Traffic 2011; 12: 703–714.
Stauber RH, Bier C, Knauer SK . Targeting taspase1 for cancer therapy–letter. Cancer Res 2012; 72: 2912.
Stumpel DJ, Schneider P, Seslija L, Osaki H, Williams O, Pieters R et al. Connectivity mapping identifies HDAC inhibitors for the treatment of t4;11-positive infant acute lymphoblastic leukemia. Leukemia 2012; 26: 682–692.
Daigle SR, Olhava EJ, Therkelsen CA, Majer CR, Sneeringer CJ, Song J et al. Selective killing of mixed lineage leukemia cells by a potent small-molecule DOT1L inhibitor. Cancer Cell 2011; 20: 53–65.
Niebuhr B, Fischer M, Täger M, Cammenga J, Stocking C . Gatekeeper function of the RUNX1 transcription factor in acute leukemia. Blood Cells Mol Dis 2008; 40: 211–218.
Wichmann C, Grez M, Lausen J . Molecular targeting of aberrant transcription factors in leukemia: strategies for RUNX1/ETO. Curr Drug Targets 2010; 11: 1181–1191.
Krämer OH, Heinzel T . Pharmacodynamic markers for histone deacetylase inhibitor development. Drug Discov Today Dis Mech 2007; 4: 277–283.
Yan M, Kanbe E, Peterson LF, Boyapati A, Miao Y, Wang Y et al. A previously unidentified alternatively spliced isoform of t8;21 transcript promotes leukemogenesis. Nat Med 2006; 12: 945–949.
Saeed S, Logie C, Stunnenberg HG, Martens JH . Genome-wide functions of PML-RARalpha in acute promyelocytic leukaemia. Br J Cancer 2011; 104: 554–558.
McCormack E, Bruserud O, Gjertsen BT . Review: genetic models of acute myeloid leukaemia. Oncogene 2008; 27: 3765–3779.
Wojiski S, Guibal FC, Kindler T, Lee BH, Jesneck JL, Fabian A et al. PML-RARalpha initiates leukemia by conferring properties of self-renewal to committed promyelocytic progenitors. Leukemia 2009; 23: 1462–1471.
Barbetti V, Gozzini A, Rovida E, Morandi A, Spinelli E, Fossati G et al. Selective anti-leukaemic activity of low-dose histone deacetylase inhibitor ITF2357 on AML1/ETO-positive cells. Oncogene 2008; 27: 1767–1778.
Tabe Y, Jin L, Contractor R, Gold D, Ruvolo P, Radke S et al. Novel role of HDAC inhibitors in AML1/ETO AML cells: activation of apoptosis and phagocytosis through induction of annexin A1. Cell Death Differ 2007; 14: 1443–1456.
Yang G, Thompson MA, Brandt SJ, Hiebert SW . Histone deacetylase inhibitors induce the degradation of the t8;21 fusion oncoprotein. Oncogene 2007; 26: 91–101.
Bug G, Schwarz K, Schoch C, Kampfmann M, Henschler R, Hoelzer D et al. Effect of histone deacetylase inhibitor valproic acid on progenitor cells of acute myeloid leukemia. Haematologica 2007; 92: 542–545.
Tabe Y, Konopleva M, Kondo Y, Contractor R, Jin L, Ruvolo V et al. PML-RARalpha and AML1-ETO translocations are rarely associated with methylation of the RARbeta2 promoter. Ann Hematol 2006; 85: 689–704.
Fanelli M, Fantozzi A, De Luca P, Caprodossi S, Matsuzawa S, Lazar MA et al. The coiled-coil domain is the structural determinant for mammalian homologues of Drosophila Sina-mediated degradation of promyelocytic leukemia protein and other tripartite motif proteins by the proteasome. J Biol Chem 2004; 279: 5374–5379.
Wichmann C, Becker Y, Chen-Wichmann L, Vogel V, Vojtkova A, Herglotz J et al. Dimer-tetramer transition controls RUNX1/ETO leukemogenic activity. Blood 2010; 116: 603–613.
Boehm J, He Y, Greiner A, Staudt L, Wirth T . Regulation of BOB.1/OBF.1 stability by SIAH. Embo J 2001; 20: 4153–4162.
Wang Y, Fiskus W, Chong DG, Buckley KM, Natarajan K, Rao R et al. Cotreatment with panobinostat and JAK2 inhibitor TG101209 attenuates JAK2V617F levels and signaling and exerts synergistic cytotoxic effects against human myeloproliferative neoplastic cells. Blood 2009; 114: 5024–5033.
Nishioka C, Ikezoe T, Yang J, Takeuchi S, Koeffler HP, Yokoyama A . MS-275, a novel histone deacetylase inhibitor with selectivity against HDAC1, induces degradation of FLT3 via inhibition of chaperone function of heat shock protein 90 in AML cells. Leuk Res 2008; 32: 1382–1392.
Lin TY, Fenger J, Murahari S, Bear MD, Kulp SK, Wang D et al. AR-42, a novel HDAC inhibitor, exhibits biologic activity against malignant mast cell lines via down-regulation of constitutively activated Kit. Blood 2010; 115: 4217–4225.
Wang L, Gural A, Sun XJ, Zhao X, Perna F, Huang G et al. The leukemogenicity of AML1-ETO is dependent on site-specific lysine acetylation. Science 2011; 333: 765–769.
Romanski A, Schwarz K, Keller M, Wietbrauk S, Vogel A, Roos J et al. Deacetylase inhibitors modulate proliferation and self-renewal properties of leukemic stem and progenitor cells. Cell Cycle 2012; 11: 17.
Corsello SM, Roti G, Ross KN, Chow KT, Galinsky I, DeAngelo DJ et al. Identification of AML1-ETO modulators by chemical genomics. Blood 2009; 113: 6193–6205.
Goto E, Tomita A, Hayakawa F, Atsumi A, Kiyoi H, Naoe T . Missense mutations in PML-RARA are critical for the lack of responsiveness to arsenic trioxide treatment. Blood 2011; 118: 1600–1609.
Kurahashi S, Hayakawa F, Miyata Y, Yasuda T, Minami Y, Tsuzuki S et al. PAX5-PML acts as a dual dominant-negative form of both PAX5 and PML. Oncogene 2011; 30: 1822–1830.
Germani A, Romero F, Houlard M, Camonis J, Gisselbrecht S, Fischer S et al. hSiah2 is a new Vav binding protein which inhibits Vav-mediated signaling pathways. Mol Cell Biol 1999; 19: 3798–3807.
Correll PH, Paulson RF, Wei X . Molecular regulation of receptor tyrosine kinases in hematopoietic malignancies. Gene 2006; 374: 26–38.
Meshinchi S, Appelbaum FR . Structural and functional alterations of FLT3 in acute myeloid leukemia. Clin Cancer Res 2009; 15: 4263–4269.
Smith CC, Wang Q, Chin CS, Salerno S, Damon LE, Levis MJ et al. Validation of ITD mutations in FLT3 as a therapeutic target in human acute myeloid leukaemia. Nature 2012; 485: 260–263.
Schmidt-Arras D, Schwäble J, Böhmer FD, Serve H . Flt3 receptor tyrosine kinase as a drug target in leukemia. Curr Pharm Des 2004; 10: 1867–1883.
Pietschmann K, Bolck HA, Buchwald M, Spielberg S, Polzer H, Spiekermann K et al. Breakdown of the FLT3-ITD/STAT5 axis and synergistic apoptosis induction by the histone deacetylase inhibitor Panobinostat and FLT3-specific inhibitors. Mol Cancer Ther; e-pub ahead of print 31 August 2012.
Weisberg E, Sattler M, Ray A, Griffin JD . Drug resistance in mutant FLT3-positive AML. Oncogene 2010; 29: 5120–5134.
Heidel F, Lipka DB, Mirea FK, Mahboobi S, Grundler R, Kancha RK et al. Bis1H-indol-2-ylmethanones are effective inhibitors of FLT3-ITD tyrosine kinase and partially overcome resistance to PKC412A in vitro. Br J Haematol 2009; 144: 865–874.
Weisberg E, Choi HG, Barrett R, Zhou W, Zhang J, Ray A et al. Discovery and characterization of novel mutant FLT3 kinase inhibitors. Mol Cancer Ther 2010; 9: 2468–2477.
Guenther MG, Lawton LN, Rozovskaia T, Frampton GM, Levine SS, Volkert TL et al. Aberrant chromatin at genes encoding stem cell regulators in human mixed-lineage leukemia. Genes Dev 2008; 22: 3403–3408.
Aveic S, Pigazzi M, Basso G . BAG1: the guardian of anti-apoptotic proteins in acute myeloid leukemia. PLoS One 2011; 6: e26097.
Inokuchi K, Yamaguchi H, Hanawa H, Tanosaki S, Nakamura K, Tarusawa M et al. Loss of DCC gene expression is of prognostic importance in acute myelogenous leukemia. Clin Cancer Res 2002; 8: 1882–1888.
Gruber FX, Ernst T, Porkka K, Engh RA, Mikkola I, Maier J et al. Dynamics of the emergence of dasatinib and nilotinib resistance in imatinib-resistant CML patients. Leukemia 2012; 26: 172–177.
Nguyen T, Dai Y, Attkisson E, Kramer L, Jordan N, Nguyen N et al. HDAC inhibitors potentiate the activity of the BCR/ABL kinase inhibitor KW-2449 in imatinib-sensitive or -resistant BCR/ABL+ leukemia cells in vitro and in vivo. Clin Cancer Res 2011; 17: 3219–3232.
Novotny-Diermayr V, Hart S, Goh KC, Cheong A, Ong LC, Hentze H et al. The oral HDAC inhibitor pracinostat SB939 is efficacious and synergistic with the JAK2 inhibitor pacritinib SB1518 in preclinical models of AML. Blood Cancer J 2012; 2: e69.
Steelman LS, Franklin RA, Abrams SL, Chappell W, Kempf CR, Basecke J et al. Roles of the Ras/Raf/MEK/ERK pathway in leukemia therapy. Leukemia 2011; 25: 1080–1094.
Martin SK, Diamond P, Gronthos S, Peet DJ, Zannettino AC . The emerging role of hypoxia, HIF-1 and HIF-2 in multiple myeloma. Leukemia 2011; 25: 1533–1542.
Cammenga J, Horn S, Bergholz U, Sommer G, Besmer P, Fiedler W et al. Extracellular KIT receptor mutants, commonly found in core binding factor AML, are constitutively active and respond to imatinib mesylate. Blood 2005; 106: 3958–3961.
Zhao HL, Ueki N, Hayman MJ . The Ski protein negatively regulates Siah2-mediated HDAC3 degradation. Biochem Biophys Res Commun 2010; 399: 623–628.
Ueki N, Zhang L, Hayman MJ . Ski can negatively regulates macrophage differentiation through its interaction with PU.1. Oncogene 2008; 27: 300–307.
Teichler S, Illmer T, Roemhild J, Ovcharenko D, Stiewe T, Neubauer A . MicroRNA29a regulates the expression of the nuclear oncogene Ski. Blood 2011; 118: 1899–1902.
Meyer PN, Fu K, Greiner TC, Smith LM, Delabie J, Gascoyne RD et al. Immunohistochemical methods for predicting cell of origin and survival in patients with diffuse large B-cell lymphoma treated with rituximab. J Clin Oncol 2011; 29: 200–207.
Haftchenary S, Avadisian M, Gunning PT . Inhibiting aberrant Stat3 function with molecular therapeutics: a progress report. Anticancer Drugs 2011; 22: 115–127.
Gupta M, Han JJ, Stenson M, Wellik L, Witzig TE . Regulation of STAT3 by histone deacetylase-3 in diffuse large B-cell lymphoma: implications for therapy. Leukemia 2012; 26: 1356–1364.
Perissi V, Jepsen K, Glass CK, Rosenfeld MG . Deconstructing repression: evolving models of co-repressor action. Nat Rev Genet 2010; 11: 109–123.
Nin DS, Kok WK, Li F, Takahashi S, Chng WJ, Khan M . Role of misfolded N-CoR mediated transcriptional deregulation of Flt3 in acute monocytic leukemia AML-M5 subtype. PLoS One 2012; 7: e34501.
Perissi V, Aggarwal A, Glass CK, Rose DW, Rosenfeld MG . A corepressor/coactivator exchange complex required for transcriptional activation by nuclear receptors and other regulated transcription factors. Cell 2004; 116: 511–526.
Ogawa S, Lozach J, Jepsen K, Sawka-Verhelle D, Perissi V, Sasik R et al. A nuclear receptor corepressor transcriptional checkpoint controlling activator protein 1-dependent gene networks required for macrophage activation. Proc Natl Acad Sci USA 2004; 101: 14461–14466.
Topol L, Jiang X, Choi H, Garrett-Beal L, Carolan PJ, Yang Y . Wnt-5a inhibits the canonical Wnt pathway by promoting GSK-3-independent beta-catenin degradation. J Cell Biol 2003; 162: 899–908.
Matsuzawa SI, Reed JC . Siah-1, SIP, and Ebi collaborate in a novel pathway for beta-catenin degradation linked to p53 responses. Mol Cell 2001; 7: 915–926.
Wang Y, Krivtsov AV, Sinha AU, North TE, Goessling W, Feng Z et al. The Wnt/beta-catenin pathway is required for the development of leukemia stem cells in AML. Science 2010; 327: 1650–1653.
Min HJ, Cho IR, Srisuttee R, Park EH, Cho DH, Ahn JH et al. Hexachlorophene suppresses beta-catenin expression by up-regulation of Siah-1 in EBV-infected B lymphoma cells. Cancer Lett 2009; 276: 136–142.
Habelhah H, Laine A, Erdjument-Bromage H, Tempst P, Gershwin ME, Bowtell DD et al. Regulation of 2-oxoglutarate alpha-ketoglutarate dehydrogenase stability by the RING finger ubiquitin ligase Siah. J Biol Chem 2004; 279: 53782–53788.
Lecuyer E, Lariviere S, Sincennes MC, Haman A, Lahlil R, Todorova M et al. Protein stability and transcription factor complex assembly determined by the SCL-LMO2 interaction. J Biol Chem 2007; 282: 33649–33658.
Makki MS, Heinzel T, Englert C . TSA downregulates Wilms tumor gene 1 Wt1 expression at multiple levels. Nucleic Acids Res 2008; 36: 4067–4078.
Hartkamp J, Roberts SG . HtrA2, taming the oncogenic activities of WT1. Cell Cycle 2010; 9: 2508–2514.
Beier UH, Akimova T, Liu Y, Wang L, Hancock WW . Histone/protein deacetylases control Foxp3 expression and the heat shock response of T-regulatory cells. Curr Opin Immunol 2011; 23: 670–678.
Ernst T, Hochhaus A . Chronic myeloid leukemia: clinical impact of BCR-ABL1 mutations and other lesions associated with disease progression. Semin Oncol 2012; 39: 58–66.
Acknowledgements
We wish to thank all members of our groups and Professor T Heinzel for helpful discussions and fruitful collaboration. Our research is supported by grants from the Deutsche Krebshilfe, the Deutsche Forschungsgemeinschaft, and the Wilhelm-Sander-Stiftung. We apologize for works not cited due to space limitations or an oversight on our part.
Author contributions
OHK performed bibliographic search, analyzed data, and wrote the paper. SKK, RHS, JH and GB commented on the article and wrote parts of the review. All authors approved the paper for publication purposes.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Gesine Bug has received honoraria and travel grants from Novartis Pharma GmbH and from Celgene GmbH. The remaining authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Krämer, O., Stauber, R., Bug, G. et al. SIAH proteins: critical roles in leukemogenesis. Leukemia 27, 792–802 (2013). https://doi.org/10.1038/leu.2012.284
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2012.284
Keywords
This article is cited by
-
Helicobacter pylori-induced gastric cancer is orchestrated by MRCKβ-mediated Siah2 phosphorylation
Journal of Biomedical Science (2021)
-
TRAF4, a new substrate of SIAH1, participates in chemotherapy resistance of breast cancer cell by counteracting SIAH1-mediated downregulation of β-catenin
Breast Cancer Research and Treatment (2020)
-
PHF19 promotes the proliferation, migration, and chemosensitivity of glioblastoma to doxorubicin through modulation of the SIAH1/β–catenin axis
Cell Death & Disease (2018)
-
Analysis of the interplay between all-trans retinoic acid and histone deacetylase inhibitors in leukemic cells
Archives of Toxicology (2017)
-
Taspase1: a 'misunderstood' protease with translational cancer relevance
Oncogene (2016)