Abstract
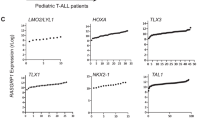
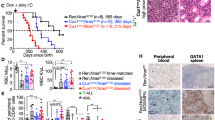
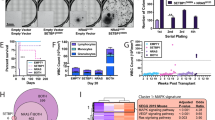
Ras guanyl nucleotide-releasing proteins (RasGRPs) are activators of Ras. Previous studies have indicated the possible involvement of RasGRP1 and RasGRP4 in leukemogenesis. Here, the predominant role of RasGRP1 in T-cell leukemogenesis is clarified. Notably, increased expression of RasGRP1, but not RasGRP4, was frequently observed in human T-cell malignancies. In a mouse bone marrow transplantation model, RasGRP1 exclusively induced T-cell acute lymphoblastic leukemia/lymphoma (T-ALL) after a shorter latency when compared with RasGRP4. Accordingly, Ba/F3 cells transduced with RasGRP1 survived longer under growth factor withdrawal or phorbol ester stimulation than those transduced with RasGRP4, presumably due to the efficient activation of Ras. Intriguingly, NOTCH1 mutations resulting in a gain of function were found in 77% of the RasGRP1-mediated mouse T-ALL samples. In addition, gain-of-function NOTCH1 mutation was found in human T-cell malignancy with elevated expression of RasGRP1. Importantly, RasGRP1 and NOTCH1 signaling cooperated in the progression of T-ALL in the murine model. The leukemogenic advantage of RasGRP1 over RasGRP4 was attenuated by the disruption of a protein kinase C phosphorylation site (RasGRP1(Thr184)) not present on RasGRP4. In conclusion, cooperation between aberrant expression of RasGRP1, a strong activator of Ras, and secondary gain-of-function mutations of NOTCH1 have an important role in T-cell leukemogenesis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
References
Aifantis I, Raetz E, Buonamici S . Molecular pathogenesis of T-cell leukaemia and lymphoma. Nat Rev Immunol 2008; 8: 380–390.
Weng AP, Ferrando AA, Lee W, Morris IV JP, Silverman LB, Sanchez-Irizarry C et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science 2004; 306: 269–271.
Chiang MY, Xu L, Shestova O, Histen G, L′heureux S, Romany C et al. Leukemia-associated NOTCH1 alleles are weak tumor initiators but accelerate K-ras-initiated leukemia. J Clin Invest 2008; 118: 3181–3194.
Dail M, Li Q, McDaniel A, Wong J, Akagi K, Huang B et al. Mutant Ikzf1, KrasG12D, and Notch1 cooperate in T lineage leukemogenesis and modulate responses to targeted agents. Proc Natl Acad Sci USA 2010; 107: 5106–5111.
Kindler T, Cornejo MG, Scholl C, Liu J, Leeman DS, Haydu JE et al. K-RasG12D-induced T-cell lymphoblastic lymphoma/leukemias harbor Notch1 mutations and are sensitive to gamma-secretase inhibitors. Blood 2008; 112: 3373–3382.
O′Neil J, Calvo J, McKenna K, Krishnamoorthy V, Aster JC, Bassing CH et al. Activating Notch1 mutations in mouse models of T-ALL. Blood 2006; 107: 781–785.
Lin YW, Nichols RA, Letterio JJ, Aplan PD . Notch1 mutations are important for leukemic transformation in murine models of precursor-T leukemia/lymphoma. Blood 2006; 107: 2540–2543.
Parikh C, Subrahmanyam R, Ren R . Oncogenic NRAS rapidly and efficiently induces CMML- and AML-like diseases in mice. Blood 2006; 108: 2349–2357.
Parikh C, Subrahmanyam R, Ren R . Oncogenic NRAS, KRAS, and HRAS exhibit different leukemogenic potentials in mice. Cancer Res 2007; 67: 7139–7146.
Wiesner SM, Jones JM, Hasz DE, Largaespada DA . Repressible transgenic model of NRAS oncogene-driven mast cell disease in the mouse. Blood 2005; 106: 1054–1062.
Chan IT, Kutok JL, Williams IR, Cohen S, Kelly L, Shigematsu H et al. Conditional expression of oncogenic K-ras from its endogenous promoter induces a myeloproliferative disease. J Clin Invest 2004; 113: 528–538.
Zhang J, Wang J, Liu Y, Sidik H, Young KH, Lodish HF et al. Oncogenic Kras-induced leukemogeneis: hematopoietic stem cells as the initial target and lineage-specific progenitors as the potential targets for final leukemic transformation. Blood 2009; 113: 1304–1314.
Van Etten RA, Shannon KM . Focus on myeloproliferative diseases and myelodysplastic syndromes. Cancer Cell 2004; 6: 547–552.
Das J, Ho M, Zikherman J, Govern C, Yang M, Weiss A et al. Digital signaling and hysteresis characterize ras activation in lymphoid cells. Cell 2009; 136: 337–351.
Forbes S, Clements J, Dawson E, Bamford S, Webb T, Dogan A et al. Cosmic 2005. Br J Cancer 2006; 94: 318–322.
Perentesis JP, Bhatia S, Boyle E, Shao Y, Shu XO, Steinbuch M et al. RAS oncogene mutations and outcome of therapy for childhood acute lymphoblastic leukemia. Leukemia 2004; 18: 685–692.
Neri A, Knowles DM, Greco A, McCormick F, Dalla-Favera R . Analysis of RAS oncogene mutations in human lymphoid malignancies. Proc Natl Acad Sci USA 1988; 8: 9268–9272.
Bowen DT, Frew ME, Hills R, Gale RE, Wheatley K, Groves MJ et al. RAS mutation in acute myeloid leukemia is associated with distinct cytogenetic subgroups but does not influence outcome in patients younger than 60 years. Blood 2005; 106: 2113–2119.
Lauchle JO, Braun BS, Loh ML, Shannon K . Inherited predispositions and hyperactive Ras in myeloid leukemogenesis. Pediatr Blood Canc 2006; 46: 579–585.
Wiemels JL, Zhang Y, Chang J, Zheng S, Metayer C, Zhang L et al. RAS mutation is associated with hyperdiploidy and parental characteristics in pediatric acute lymphoblastic leukemia. Leukemia 2005; 19: 415–419.
Graux C, Cools J, Michaux L, Vandenberghe P, Hagemeijer A . Cytogenetics and molecular genetics of T-cell acute lymphoblastic leukemia: from thymocyte to lymphoblast. Leukemia 2006; 20: 1496–1510.
Coughlin JJ, Stang SL, Dower NA, Stone JC . The role of RasGRPs in regulation of lymphocyte proliferation. Immunol Lett 2006; 105: 77–82.
Stone JC . Regulation of Ras in lymphocytes: get a GRP. Biochem Soc Trans 2006; 34: 858–861.
Zheng Y, Liu H, Coughlin J, Zheng J, Li L, Stone JC . Phosphorylation of RasGRP3 on threonine 133 provides a mechanistic link between PKC and Ras signaling systems in B cells. Blood 2005; 105: 3648–3654.
Aiba Y, Oh-hora M, Kiyonaka S, Kimura Y, Hijikata A, Mori Y et al. Activation of RasGRP3 by phosphorylation of Thr-133 is required for B cell receptor-mediated Ras activation. Proc Natl Acad Sci USA 2004; 101: 16612–16617.
Dower NA, Stang SL, Bottorff DA, Ebinu JO, Dickie P, Ostergaard HL et al. RasGRP is essential for mouse thymocyte differentiation and TCR signaling. Nat Immunol 2000; 1: 317–321.
Layer K, Lin G, Nencioni A, Hu W, Schmucker A, Antov AN et al. Autoimmunity as the consequence of a spontaneous mutation in Rasgrp1. Immunity 2003; 19: 243–255.
Beaulieu N, Zahedi B, Goulding RE, Tazmini G, Anthony KV, Omeis SL et al. Regulation of RasGRP1 by B cell antigen receptor requires cooperativity between three domains controlling translocation to the plasma membrane. Mol Biol Cell 2007; 18: 3156–3168.
Coughlin JJ, Stang SL, Dower NA, Stone JC . RasGRP1 and RasGRP3 regulate B cell proliferation by facilitating B cell receptor-Ras signaling. J Immunol 2005; 175: 7179–7184.
Liu Y, Zhu M, Nishida K, Hirano T, Zhang W . An essential role for RasGRP1 in mast cell function and IgE-mediated allergic response. J Exp Med 2007; 204: 93–103.
Yang Y, Li L, Wong GW, Krilis SA, Madhusudhan MS, Sali A et al. RasGRP4, a new mast cell-restricted Ras guanine nucleotide-releasing protein with calcium- and diacylglycerol-binding motifs. Identification of defective variants of this signaling protein in asthma, mastocytosis, and mast cell leukemia patients and demonstration of the importance of RasGRP4 in mast cell development and function. J Biol Chem 2002; 277: 25756–25774.
Li L, Yang Y, Wong GW, Stevens RL . Mast cells in airway hyporesponsive C3H/HeJ mice express a unique isoform of the signaling protein Ras guanine nucleotide releasing protein 4 that is unresponsive to diacylglycerol and phorbol esters. J Immunol 2003; 171: 390–397.
Reuther GW, Lambert QT, Rebhun JF, Caligiuri MA, Quilliam LA, Der CJ . RasGRP4 is a novel Ras activator isolated from acute myeloid leukemia. J Biol Chem 2002; 277: 30508–30514.
Watanabe-Okochi N, Oki T, Komeno Y, Kato N, Yuji K, Ono R et al. Possible involvement of RasGRP4 in leukemogenesis. Int J Hematol 2009; 89: 470–481.
Klinger MB, Guilbault B, Goulding RE, Kay RJ . Deregulated expression of RasGRP1 initiates thymic lymphomagenesis independently of T-cell receptors. Oncogene 2005; 24: 2695–2704.
Norment AM, Bogatzki LY, Klinger M, Ojala EW, Bevan MJ, Kay RJ . Transgenic expression of RasGRP1 induces the maturation of double-negative thymocytes and enhances the production of CD8 single-positive thymocytes. J Immunol 2003; 170: 1141–1149.
Oki-Idouchi CE, Lorenzo PS . Transgenic overexpression of RasGRP1 in mouse epidermis results in spontaneous tumors of the skin. Cancer Res 2007; 67: 276–280.
Mikkers H, Allen J, Knipscheer P, Romeijn L, Hart A, Vink E et al. High-throughput retroviral tagging to identify components of specific signaling pathways in cancer. Nat Genet 2002; 32: 153–159.
Raponi M, Lancet JE, Fan H, Dossey L, Lee G, Gojo I et al. A 2-gene classifier for predicting response to the farnesyltransferase inhibitor tipifarnib in acute myeloid leukemia. Blood 2008; 111: 2589–2596.
Ding H, Hackbarth J, Schneider PA, Peterson KL, Meng XW, Dai H et al. Cytotoxicity of farnesyltransferase inhibitors in lymphoid cells mediated by MAPK pathway inhibition and Bim upregulation. Blood 2011; 118: 4872–4881.
Lauchle JO, Kim D, Le DT, Akagi K, Crone M, Krisman K et al. Response and resistance to MEK inhibition in leukaemias initiated by hyperactive Ras. Nature 2009; 461: 411–414.
Ding Y, Harada Y, Imagawa J, Kimura A, Harada H . AML1/RUNX1 point mutation possibly promotes leukemic transformation in myeloproliferative neoplasms. Blood 2009; 114: 5201–5205.
Kitamura T, Koshino Y, Shibata F, Oki T, Nakajima H, Nosaka T et al. Retrovirus-mediated gene transfer and expression cloning: powerful tools in functional genomics. Exp Hematol 2003; 31: 1007–1014.
Morita S, Kojima T, Kitamura T . Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther 2000; 7: 1063–1066.
Lu Y, Kitaura J, Oki T, Komeno Y, Ozaki K, Kiyono M et al. Identification of TSC-22 as a potential tumor suppressor that is upregulated by Flt3-D835V but not Flt3-ITD. Leukemia 2007; 21: 2246–2257.
Kato N, Kitaura J, Doki N, Komeno Y, Watanabe-Okochi N, Togami K et al. Two types of C/EBPα mutations play distinct but collaborative roles in leukemogenesis: lessons from clinical data and BMT models. Blood 2011; 117: 221–233.
Nakahara F, Sakata-Yanagimoto M, Komeno Y, Kato N, Uchida T, Haraguchi K et al. Hes1 immortalizes committed progenitors and plays a role in blast crisis transition in chronic myelogenous leukemia. Blood 2010; 115: 2872–2881.
Yeoh EJ, Ross ME, Shurtleff SA, Williams WK, Patel D, Mahfouz R et al. Classification, subtype discovery, and prediction of outcome in pediatric acute lymphoblastic leukemia by gene expression profiling. Cancer Cell 2002; 1: 133–143.
Asnafi V, Beldjord K, Boulanger E, Comba B, Le Tutour P, Estienne MH et al. Analysis of TCR, pT alpha, and RAG-1 in T-acute lymphoblastic leukemias improves understanding of early human T-lymphoid lineage commitment. Blood 2003; 101: 2693–2703.
Balgobind BV, Van Vlierberghe P, van den Ouweland AM, Beverloo HB, Terlouw-Kromosoeto JN, van Wering ER et al. Leukemia-associated NF1 inactivation in patients with pediatric T-ALL and AML lacking evidence for neurofibromatosis. Blood 2008; 111: 4322–4328.
Acknowledgements
We thank Dr Shigeru Chiba for kindly providing plasmids. This work was supported by the Ministry of Education, Science, Technology, Sports and Culture, Japan.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
TK serves as a consultant for R&D Systems. The other authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Leukemia website
Rights and permissions
About this article
Cite this article
Oki, T., Kitaura, J., Watanabe-Okochi, N. et al. Aberrant expression of RasGRP1 cooperates with gain-of-function NOTCH1 mutations in T-cell leukemogenesis. Leukemia 26, 1038–1045 (2012). https://doi.org/10.1038/leu.2011.328
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2011.328
Keywords
This article is cited by
-
Identification of leukemia stem cell expression signatures through Monte Carlo feature selection strategy and support vector machine
Cancer Gene Therapy (2020)
-
Increased baseline RASGRP1 signals enhance stem cell fitness during native hematopoiesis
Oncogene (2020)
-
RasGRP1 overexpression in T-ALL increases basal nucleotide exchange on Ras rendering the Ras/PI3K/Akt pathway responsive to protumorigenic cytokines
Oncogene (2016)
-
MEK and PI3K-AKT inhibitors synergistically block activated IL7 receptor signaling in T-cell acute lymphoblastic leukemia
Leukemia (2016)
-
Dysregulated choline metabolism in T-cell lymphoma: role of choline kinase-α and therapeutic targeting
Blood Cancer Journal (2015)