Abstract
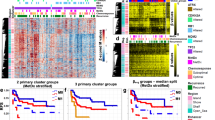
This study was designed to analyze the effect of global and gene-specific DNA methylation patterns on the outcome of patients with acute myeloid leukemia (AML). Methylation of CDKN2B (p15), E-cadherin (CDH) and hypermethylated in cancer 1 (HIC1) promoters and global DNA methylation by luminometric methylation assay (LUMA) was analyzed in 107 AML patients and cytogenetic and molecular mutational analysis was performed. In addition, genome-wide promoter-associated methylation was assessed using the Illumina HumanMethylation27 array in a proportion of the patients. Promoter methylation was discovered in 66, 66 and 51% of the patients for p15, CDH and HIC1, respectively. In multivariate analysis, low global DNA methylation was associated with higher complete remission rate (hazard ratio (HR) 5.9, P=0.005) and p15 methylation was associated with better overall (HR 0.4, P=0.001) and disease-free survival (HR 0.4, P=0.016). CDH and HIC1 methylation were not associated with clinical outcome. Mutational status and karyotype were not significantly associated with gene-specific methylation or global methylation. Increased genome-wide promoter-associated methylation was associated with better overall and disease-free survival as well as with LUMA hypomethylation. We conclude that global and gene-specific methylation patterns are independently associated with the clinical outcome in AML patients.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Frohling S, Scholl C, Gilliland DG, Levine RL . Genetics of myeloid malignancies: pathogenetic and clinical implications. J Clin Oncol 2005; 23: 6285–6295.
Rowley JD . The critical role of chromosome translocations in human leukemias. Annu Rev Genet 1998; 32: 495–519.
Feinberg AP, Ohlsson R, Henikoff S . The epigenetic progenitor origin of human cancer. Nat Rev Genet 2006; 7: 21–33.
Feinberg AP, Vogelstein B . Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature 1983; 301: 89–92.
Melki JR, Vincent PC, Clark SJ . Concurrent DNA hypermethylation of multiple genes in acute myeloid leukemia. Cancer Res 1999; 59: 3730–3740.
Li Q, Kopecky KJ, Mohan A, Willman CL, Appelbaum FR, Weick JK et al. Estrogen receptor methylation is associated with improved survival in adult acute myeloid leukemia. Clin Cancer Res 1999; 5: 1077–1084.
Chim CS, Tam CY, Liang R, Kwong YL . Methylation of p15 and p16 genes in adult acute leukemia: lack of prognostic significance. Cancer 2001; 91: 2222–2229.
Galm O, Wilop S, Luders C, Jost E, Gehbauer G, Herman JG et al. Clinical implications of aberrant DNA methylation patterns in acute myelogenous leukemia. Ann Hematol 2005; 84 (Suppl 1): 39–46.
Shimamoto T, Ohyashiki JH, Ohyashiki K . Methylation of p15(INK4b) and E-cadherin genes is independently correlated with poor prognosis in acute myeloid leukemia. Leuk Res 2005; 29: 653–659.
Grovdal M, Khan R, Aggerholm A, Antunovic P, Astermark J, Bernell P et al. Negative effect of DNA hypermethylation on the outcome of intensive chemotherapy in older patients with high-risk myelodysplastic syndromes and acute myeloid leukemia following myelodysplastic syndrome. Clin Cancer Res 2007; 13: 7107–7112.
Vardiman JW, Harris NL, Brunning RD . The World Health Organization (WHO) classification of the myeloid neoplasms. Blood 2002; 100: 2292–2302.
Schlenk RF, Dohner K, Krauter J, Frohling S, Corbacioglu A, Bullinger L et al. Mutations and treatment outcome in cytogenetically normal acute myeloid leukemia. N Engl J Med 2008; 358: 1909–1918.
Zeschnigk M, Lich C, Buiting K, Doerfler W, Horsthemke B . A single-tube PCR test for the diagnosis of Angelman and Prader-Willi syndrome based on allelic methylation differences at the SNRPN locus. Eur J Hum Genet 1997; 5: 94–98.
Aggerholm A, Holm MS, Guldberg P, Olesen LH, Hokland P . Promoter hypermethylation of p15INK4B, HIC1, CDH1, and ER is frequent in myelodysplastic syndrome and predicts poor prognosis in early-stage patients. Eur J Haematol 2006; 76: 23–32.
Cremonesi L, Firpo S, Ferrari M, Righetti PG, Gelfi C . Double-gradient DGGE for optimized detection of DNA point mutations. Biotechniques 1997; 22: 326–330.
Geli J, Kiss N, Karimi M, Lee JJ, Backdahl M, Ekstrom TJ et al. Global and regional CpG methylation in pheochromocytomas and abdominal paragangliomas: association to malignant behavior. Clin Cancer Res 2008; 14: 2551–2559.
Guldberg P, Worm J, Gronbaek K . Profiling DNA methylation by melting analysis. Methods 2002; 27: 121–127.
Karimi M, Johansson S, Stach D, Corcoran M, Grander D, Schalling M et al. LUMA (LUminometric Methylation Assay)--a high throughput method to the analysis of genomic DNA methylation. Exp Cell Res 2006; 312: 1989–1995.
Karimi M, Johansson S, Ekstrom TJ . Using LUMA: a Luminometric-based assay for global DNA-methylation. Epigenetics 2006; 1: 45–48.
Fan JB, Oliphant A, Shen R, Kermani BG, Garcia F, Gunderson KL et al. Highly parallel SNP genotyping. Cold Spring Harb Symp Quant Biol 2003; 68: 69–78.
Fan JB, Gunderson KL, Bibikova M, Yeakley JM, Chen J, Wickham Garcia E et al. Illumina universal bead arrays. Methods Enzymol 2006; 410: 57–73.
Wong IH, Ng MH, Huang DP, Lee JC . Aberrant p15 promoter methylation in adult and childhood acute leukemias of nearly all morphologic subtypes: potential prognostic implications. Blood 2000; 95: 1942–1949.
Issa JP . CpG island methylator phenotype in cancer. Nat Rev Cancer 2004; 4: 988–993.
Roman-Gomez J, Jimenez-Velasco A, Agirre X, Castillejo JA, Navarro G, Garate L et al. Promoter hypermethylation and global hypomethylation are independent epigenetic events in lymphoid leukemogenesis with opposing effects on clinical outcome. Leukemia 2006; 20: 1445–1448.
Soares J, Pinto AE, Cunha CV, Andre S, Barao I, Sousa JM et al. Global DNA hypomethylation in breast carcinoma: correlation with prognostic factors and tumor progression. Cancer 1999; 85: 112–118.
Kroeger H, Jelinek J, Estecio MR, He R, Kondo K, Chung W et al. Aberrant CpG island methylation in acute myeloid leukemia is accentuated at relapse. Blood 2008; 112: 1366–1373.
Costello JF, Plass C . Methylation matters. J Med Genet 2001; 38: 285–303.
Shang D, Liu Y, Matsui Y, Ito N, Nishiyama H, Kamoto T et al. Demethylating agent 5-aza-2′-deoxycytidine enhances susceptibility of bladder transitional cell carcinoma to Cisplatin. Urology 2008; 71: 1220–1225.
Festuccia C, Gravina GL, D’Alessandro AM, Muzi P, Millimaggi D, Dolo V et al. Azacitidine improves antitumor effects of docetaxel and cisplatin in aggressive prostate cancer models. Endocr Relat Cancer 2009; 16: 401–413.
Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci USA 2005; 102: 10604–10609.
Bjornsson HT, Sigurdsson MI, Fallin MD, Irizarry RA, Aspelund T, Cui H et al. Intra-individual change over time in DNA methylation with familial clustering. JAMA 2008; 299: 2877–2883.
Fazzari MJ, Greally JM . Epigenomics: beyond CpG islands. Nat Rev Genet 2004; 5: 446–455.
Romermann D, Hasemeier B, Metzig K, Gohring G, Schlegelberger B, Langer F et al. Global increase in DNA methylation in patients with myelodysplastic syndrome. Leukemia 2008; 22: 1954–1956.
Esteller M . Epigenetics in cancer. N Engl J Med 2008; 358: 1148–1159.
Feinberg AP, Tycko B . The history of cancer epigenetics. Nat Rev Cancer 2004; 4: 143–153.
Olesen LH, Aggerholm A, Andersen BL, Nyvold CG, Guldberg P, Norgaard JM et al. Molecular typing of adult acute myeloid leukaemia: significance of translocations, tandem duplications, methylation, and selective gene expression profiling. Br J Haematol 2005; 131: 457–467.
Kroeger H, Jelinek J, Komblau SM, Bueso-Ramos CE, Issa JP . Increased DNA methylation is associated with good prognosis in AML. 49th Annual Meeting of the American-Society-of-Hematology, 2007, Atlanta, GA, 2007, p 595.
Ogino S, Nosho K, Kirkner GJ, Kawasaki T, Meyerhardt JA, Loda M et al. CpG island methylator phenotype, microsatellite instability, BRAF mutation and clinical outcome in colon cancer. Gut 2009; 58: 90–96.
Figueroa ME, Skrabanek L, Li Y, Jiemjit A, Fandy TE, Paietta E et al. MDS and secondary AML display unique patterns and abundance of aberrant DNA methylation. Blood 2009; 114: 3363–3364.
Jiang Y, Dunbar A, Gondek LP, Mohan S, Rataul M, O’Keefe C et al. Aberrant DNA methylation is a dominant mechanism in MDS progression to AML. Blood 2009; 113: 1315–1325.
Deneberg S, Grovdall M, Jansson M, Gaidzik VI, Corbacioglu A, Nahi H et al. Different incidence and implications of DNA hypermethylation in de novo AML compared to high-risk MDS and AML following MDS. 50th Annual Meeting of the American- Society-of-Hematology, 2008, 06–09 December, San Francisco, CA, 2008, p 1146–1147.
Acknowledgements
This study was supported by the Swedish Cancer Foundation, Swedish Research Council and Stockholm County Council. MK was supported by a KI post doc grant.
A special thanks to Professor Per Guldberg in Copenhagen for his hospitality and help regarding methylation assay methodology.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Authorship
Stefan Deneberg collected clinical data, analyzed data and wrote the article. Michael Grövdal performed the DGGE analyses and co-wrote the article. Mohsen Karimi performed the LUMA, MS-MCA and pyrosequencing. Monika Jansson performed DGGE analyses. Hareth Nahi helped collect clinical information. Andrea Corbacioglu, Verena Gaidzik and Konstanze Döhner performed the molecular analyses. Christer Paul co-wrote the article and supervised sample collection and storage. Tomas J Ekström performed the LUMA analysis and co-wrote the article. Eva Hellström-Lindberg helped design the study, assisted in data analysis and co-wrote the article. Sören Lehmann designed the study and co-wrote the article.
Supplementary Information accompanies the paper on the Leukemia website
Rights and permissions
About this article
Cite this article
Deneberg, S., Grövdal, M., Karimi, M. et al. Gene-specific and global methylation patterns predict outcome in patients with acute myeloid leukemia. Leukemia 24, 932–941 (2010). https://doi.org/10.1038/leu.2010.41
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2010.41
Keywords
This article is cited by
-
DNA methylation and cancer incidence: lymphatic–hematopoietic versus solid cancers in the Strong Heart Study
Clinical Epigenetics (2021)
-
A vicious loop of fatty acid-binding protein 4 and DNA methyltransferase 1 promotes acute myeloid leukemia and acts as a therapeutic target
Leukemia (2018)
-
Clinical implications of genome-wide DNA methylation studies in acute myeloid leukemia
Journal of Hematology & Oncology (2017)
-
DNA methylation modifies the association between obesity and survival after breast cancer diagnosis
Breast Cancer Research and Treatment (2016)
-
HIC1 modulates uveal melanoma progression by activating lncRNA-numb
Tumor Biology (2016)