Abstract
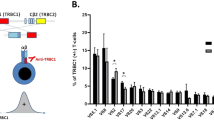
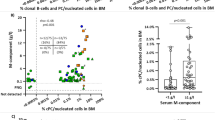
Monoclonal B-cell lymphocytosis (MBL) is a preclinical hematologic syndrome characterized by small accumulations of CD5+ B lymphocytes. Most MBL share phenotypic characteristics with chronic lymphocytic leukemia (CLL). Although some MBL progress to CLL, most MBL have apparently limited potential for progression to CLL, particularly those MBL with normal absolute B-cell counts (‘low-count’ MBL). Most CLL are monoclonal and it is not known whether MBL are monoclonal or oligoclonal; this is important because it is unclear whether MBL represent indolent CLL or represent a distinct premalignant precursor before the development of CLL. We used flow cytometry analysis and sorting to determine immunophenotypic characteristics, clonality and molecular features of MBL from familial CLL kindreds. Single-cell analysis indicated four of six low-count MBL consisted of two or more unrelated clones; the other two MBL were monoclonal. 87% of low-count MBL clones had mutated immunoglobulin genes, and no immunoglobulin heavy-chain rearrangements of VH family 1 were observed. Some MBL were diversified, clonally related populations with evidence of antigen drive. We conclude that although low-count MBL share many phenotypic characteristics with CLL, many MBL are oligoclonal. This supports a model for step-wise development of MBL into CLL.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Chiorazzi N, Rai KR, Ferrarini M . Chronic lymphocytic leukemia. N Engl J Med 2005; 352: 804–815.
Dighiero G, Hamblin TJ . Chronic lymphocytic leukaemia. Lancet 2008; 371: 1017–1029.
Goldin LR, Bjorkholm M, Kristinsson SY, Turesson I, Landgren O . Elevated risk of chronic lymphocytic leukemia and other indolent non-Hodgkin's lymphomas among relatives of patients with chronic lymphocytic leukemia. Haematologica 2009; 94: 647–653.
Linet MS, Schubauer-Berigan MK, Weisenburger DD, Richardson DB, Landgren O, Blair A et al. Chronic lymphocytic leukaemia: an overview of aetiology in light of recent developments in classification and pathogenesis. Br J Haematol 2007; 139: 672–686.
Fischer M, Klein U, Kuppers R . Molecular single-cell analysis reveals that CD5-positive peripheral blood B cells in healthy humans are characterized by rearranged Vkappa genes lacking somatic mutation. J Clin Invest 1997; 100: 1667–1676.
Geiger KD, Klein U, Brauninger A, Berger S, Leder K, Rajewsky K et al. CD5-positive B cells in healthy elderly humans are a polyclonal B cell population. Eur J Immunol 2000; 30: 2918–2923.
Klein U, Rajewsky K, Kuppers R . Human immunoglobulin (Ig)M+IgD+ peripheral blood B cells expressing the CD27 cell surface antigen carry somatically mutated variable region genes: CD27 as a general marker for somatically mutated (memory) B cells. J Exp Med 1998; 188: 1679–1689.
Fais F, Ghiotto F, Hashimoto S, Sellars B, Valetto A, Allen SL et al. Chronic lymphocytic leukemia B cells express restricted sets of mutated and unmutated antigen receptors. J Clin Invest 1998; 102: 1515–1525.
Weinberg JB, Volkheimer AD, Chen Y, Beasley BE, Jiang N, Lanasa MC et al. Clinical and molecular predictors of disease severity and survival in chronic lymphocytic leukemia. Am J Hematol 2007; 82: 1063–1070.
Hallek M, Cheson BD, Catovsky D, Caligaris-Cappio F, Dighiero G, Dohner H et al. Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: a report from the International Workshop on Chronic Lymphocytic Leukemia updating the National Cancer Institute–Working Group 1996 guidelines. Blood 2008; 111: 5446–5456.
Marti GE, Rawstron AC, Ghia P, Hillmen P, Houlston RS, Kay N et al. Diagnostic criteria for monoclonal B-cell lymphocytosis. Br J Haematol 2005; 130: 325–332.
Marti G, Abbasi F, Raveche E, Rawstron AC, Ghia P, Aurran T et al. Overview of monoclonal B-cell lymphocytosis. Br J Haematol 2007; 139: 701–708.
Ghia P, Prato G, Scielzo C, Stella S, Geuna M, Guida G et al. Monoclonal CD5+ and CD5- B-lymphocyte expansions are frequent in the peripheral blood of the elderly. Blood 2004; 103: 2337–2342.
Rawstron AC, Green MJ, Kuzmicki A, Kennedy B, Fenton JA, Evans PA et al. Monoclonal B lymphocytes with the characteristics of ‘indolent’ chronic lymphocytic leukemia are present in 3.5% of adults with normal blood counts. Blood 2002; 100: 635–639.
Nieto WG, Almeida J, Romero A, Teodosio C, Lopez A, Henriques AF et al. Increased frequency (12%) of circulating chronic lymphocytic leukemia-like B-cell clones in healthy subjects using a highly sensitive multicolor flow cytometry approach. Blood 2009; 114: 33–37.
Rawstron AC, Yuille MR, Fuller J, Cullen M, Kennedy B, Richards SJ et al. Inherited predisposition to CLL is detectable as subclinical monoclonal B-lymphocyte expansion. Blood 2002; 100: 2289–2290.
Marti GE, Carter P, Abbasi F, Washington GC, Jain N, Zenger VE et al. B-cell monoclonal lymphocytosis and B-cell abnormalities in the setting of familial B-cell chronic lymphocytic leukemia. Cytometry B Clin Cytom 2003; 52: 1–12.
Rawstron AC, Bennett FL, O’Connor SJ, Kwok M, Fenton JA, Plummer M et al. Monoclonal B-cell lymphocytosis and chronic lymphocytic leukemia. N Engl J Med 2008; 359: 575–583.
Landgren O, Albitar M, Ma W, Abbasi F, Hayes RB, Ghia P et al. B-cell clones as early markers for chronic lymphocytic leukemia. N Engl J Med 2009; 360: 659–667.
Dagklis A, Fazi C, Sala C, Cantarelli V, Scielzo C, Massacane R et al. The immunoglobulin gene repertoire of low-count chronic lymphocytic leukemia (CLL)-like monoclonal B lymphocytosis is different from CLL: diagnostic implications for clinical monitoring. Blood 2009; 114: 26–32.
Cheson BD, Bennett JM, Grever M, Kay N, Keating MJ, O’Brien S et al. National Cancer Institute-sponsored Working Group guidelines for chronic lymphocytic leukemia: revised guidelines for diagnosis and treatment. Blood 1996; 87: 4990–4997.
Volkheimer AD, Weinberg JB, Beasley BE, Whitesides JF, Gockerman JP, Moore JO et al. Progressive immunoglobulin gene mutations in chronic lymphocytic leukemia: evidence for antigen-driven intraclonal diversification. Blood 2007; 109: 1559–1567.
Lefranc MP, Giudicelli V, Kaas Q, Duprat E, Jabado-Michaloud J, Scaviner D et al. IMGT, the international ImMunoGeneTics information system. Nucleic Acids Res 2005; 33 (Database issue): D593–D597.
Lossos IS, Tibshirani R, Narasimhan B, Levy R . The inference of antigen selection on Ig genes. J Immunol 2000; 165: 5122–5126.
Gurrieri C, McGuire P, Zan H, Yan XJ, Cerutti A, Albesiano E et al. Chronic lymphocytic leukemia B cells can undergo somatic hypermutation and intraclonal immunoglobulin V(H)DJ(H) gene diversification. J Exp Med 2002; 196: 629–639.
Abbasi F, Longo NS, Lipsky PE, Raveche E, Schleinitz TA, Stetler-Stevenson M et al. B-cell repertoire and clonal analysis in unaffected first degree relatives in familial chronic lymphocytic leukaemia kindred. Br J Haematol 2007; 139: 820–823.
Rawstron AC, Bennett F, Hillmen P . The biological and clinical relationship between CD5+23+ monoclonal B-cell lymphocytosis and chronic lymphocytic leukaemia. Br J Haematol 2007; 139: 724–729.
Degan M, Bomben R, Bo MD, Zucchetto A, Nanni P, Rupolo M et al. Analysis of IgV gene mutations in B cell chronic lymphocytic leukaemia according to antigen-driven selection identifies subgroups with different prognosis and usage of the canonical somatic hypermutation machinery. Br J Haematol 2004; 126: 29–42.
Efremov DG, Ivanovski M, Batista FD, Pozzato G, Burrone OR . IgM-producing chronic lymphocytic leukemia cells undergo immunoglobulin isotype-switching without acquiring somatic mutations. J Clin Invest 1996; 98: 290–298.
Dohner H, Stilgenbauer S, Benner A, Leupolt E, Krober A, Bullinger L et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med 2000; 343: 1910–1916.
Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA 2002; 99: 15524–15529.
Fung SS, Hillier KL, Leger CS, Sandhu I, Vickars LM, Galbraith PF et al. Clinical progression and outcome of patients with monoclonal B-cell lymphocytosis. Leuk Lymphoma 2007; 48: 1087–1091.
Shanafelt TD, Kay NE, Call TG, Zent CS, Jelinek DF, LaPlant B et al. MBL or CLL: which classification best categorizes the clinical course of patients with an absolute lymphocyte count >or=5 × 10(9) L(−1) but a B-cell lymphocyte count Leuk Res 2008; 32: 1458–1461.
Chang H, Cerny J . Molecular characterization of chronic lymphocytic leukemia with two distinct cell populations: evidence for separate clonal origins. Am J Clin Pathol 2006; 126: 23–28.
Herve M, Xu K, Ng YS, Wardemann H, Albesiano E, Messmer BT et al. Unmutated and mutated chronic lymphocytic leukemias derive from self-reactive B cell precursors despite expressing different antibody reactivity. J Clin Invest 2005; 115: 1636–1643.
Ghia P, Stamatopoulos K, Belessi C, Moreno C, Stella S, Guida G et al. Geographic patterns and pathogenetic implications of IGHV gene usage in chronic lymphocytic leukemia: the lesson of the IGHV3-21 gene. Blood 2005; 105: 1678–1685.
Murray F, Darzentas N, Hadzidimitriou A, Tobin G, Boudjogra M, Scielzo C et al. Stereotyped patterns of somatic hypermutation in subsets of patients with chronic lymphocytic leukemia: implications for the role of antigen selection in leukemogenesis. Blood 2008; 111: 1524–1533.
Brezinschek HP, Foster SJ, Brezinschek RI, Dorner T, Domiati-Saad R, Lipsky PE . Analysis of the human VH gene repertoire. Differential effects of selection and somatic hypermutation on human peripheral CD5(+)/IgM+ and CD5(−)/IgM+ B cells. J Clin Invest 1997; 99: 2488–2501.
Krober A, Seiler T, Benner A, Bullinger L, Bruckle E, Lichter P et al. V(H) mutation status, CD38 expression level, genomic aberrations, and survival in chronic lymphocytic leukemia. Blood 2002; 100: 1410–1416.
Ng D, Toure O, Wei MH, Arthur DC, Abbasi F, Fontaine L et al. Identification of a novel chromosome region, 13q21.33–q22.2, for susceptibility genes in familial chronic lymphocytic leukemia. Blood 2007; 109: 916–925.
Acknowledgements
We thank Dr Andy Rawstron for his thoughtful review of this paper. We thank the study participants for their willingness to participate in this study, and the hematology-oncology physicians, nurses and physician assistants for their special help. Flow cytometry was performed in the Duke Human Vaccine Institute Flow Cytometry Core Facility that is supported by the National Institutes of Health award AI-51445.
Grant Support: MC Lanasa is a fellow of the Leukemia and Lymphoma Society of America. This research was funded by the Bernstein Fund for Leukemia Research, the VA Research Service and a grant from the National Institutes of Health (NCI R03 CA128030).
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on the Leukemia website (http://www.nature.com/leu)
Supplementary information
Rights and permissions
About this article
Cite this article
Lanasa, M., Allgood, S., Volkheimer, A. et al. Single-cell analysis reveals oligoclonality among ‘low-count’ monoclonal B-cell lymphocytosis. Leukemia 24, 133–140 (2010). https://doi.org/10.1038/leu.2009.192
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2009.192
Keywords
This article is cited by
-
Pathophysiology of chronic lymphocytic leukemia and human B1 cell development
International Journal of Hematology (2020)
-
Multiple productive IGH rearrangements denote oligoclonality even in immunophenotypically monoclonal CLL
Leukemia (2018)
-
Lymphoma-like monoclonal B cell lymphocytosis in a patient population: biology, natural evolution, and differences from CLL-like clones
Annals of Hematology (2018)
-
Addressing heterogeneity of individual blood cancers: the need for single cell analysis
Cell Biology and Toxicology (2017)
-
Next-generation IgVH sequencing CLL-like monoclonal B-cell lymphocytosis reveals frequent oligoclonality and ongoing hypermutation
Leukemia (2016)