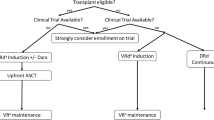
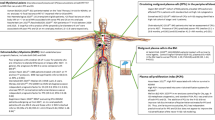
Abstract
Myeloma is a malignant proliferation of monoclonal plasma cells. Although morphologically similar, several subtypes of the disease have been identified at the genetic and molecular level. These genetic subtypes are associated with unique clinicopathological features and dissimilar outcome. At the top hierarchical level, myeloma can be divided into hyperdiploid and non-hyperdiploid subtypes. The latter is mainly composed of cases harboring IgH translocations, generally associated with more aggressive clinical features and shorter survival. The three main IgH translocations in myeloma are the t(11;14)(q13;q32), t(4;14)(p16;q32) and t(14;16)(q32;q23). Trisomies and a more indolent form of the disease characterize hyperdiploid myeloma. A number of genetic progression factors have been identified including deletions of chromosomes 13 and 17 and abnormalities of chromosome 1 (1p deletion and 1q amplification). Other key drivers of cell survival and proliferation have also been identified such as nuclear factor- B-activating mutations and other deregulation factors for the cyclin-dependent pathways regulators. Further understanding of the biological subtypes of the disease has come from the application of novel techniques such as gene expression profiling and array-based comparative genomic hybridization. The combination of data arising from these studies and that previously elucidated through other mechanisms allows for most myeloma cases to be classified under one of several genetic subtypes. This paper proposes a framework for the classification of myeloma subtypes and provides recommendations for genetic testing. This group proposes that genetic testing needs to be incorporated into daily clinical practice and also as an essential component of all ongoing and future clinical trials.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Kyle RA, Rajkumar SV . Multiple myeloma [see comment]. [published erratum appears in N Engl J Med 2005; 352:1163] N Engl J Med 2004; 351: 1860–1873.
Fonseca R . Many and multiple myeloma(s). Leukemia 2003; 17: 1943–1944.
Zhan F, Huang Y, Colla S, Stewart JP, Hanamura I, Gupta S et al. The molecular classification of multiple myeloma. Blood 2006; 108: 2020–2028.
Ludwig H, Durie BG, Bolejack V, Turesson I, Kyle RA, Blade J et al. Myeloma in patients younger than age 50 years presents with more favorable features and shows better survival: an analysis of 10 549 patients from the International Myeloma Working Group. Blood 2008; 111: 4039–4047.
Bergsagel PL, Kuehl WM, Zhan F, Sawyer J, Barlogie B, Shaughnessy J et al. Cyclin D dysregulation: an early and unifying pathogenic event in multiple myeloma. Blood 2005; 106: 296–303.
Avet-Loiseau H, Attal M, Moreau P, Charbonnel C, Garban F, Hulin C et al. Genetic abnormalities and survival in multiple myeloma: the experience of the Intergroupe Francophone du Myelome. Blood 2007; 109: 3489–3495.
Fonseca R, Blood E, Rue M, Harrington D, Oken MM, Kyle RA et al. Clinical and biologic implications of recurrent genomic aberrations in myeloma. Blood 2003; 101: 4569–4575.
Debes-Marun C, Dewald G, Bryant S, Picken E, Santana-Dávila S, González-Paz N et al. Chromosome abnormalities clustering and its implications for pathogenesis and prognosis in myeloma. Leukemia 2003; 17: 427–436.
Smadja NV, Fruchart C, Isnard F, Louvet C, Dutel JL, Cheron N et al. Chromosomal analysis in multiple myeloma: cytogenetic evidence of two different diseases. Leukemia 1998; 12: 960–969.
Carrasco DR, Tonon G, Huang Y, Zhang Y, Sinha R, Feng B et al. High-resolution genomic profiles define distinct clinico-pathogenetic subgroups of multiple myeloma patients. Cancer Cell 2006; 9: 313–325.
Keats JJ, Reiman T, Maxwell CA, Taylor BJ, Larratt LM, Mant MJ et al. In multiple myeloma, t(4;14)(p16;q32) is an adverse prognostic factor irrespective of FGFR3 expression. Blood 2003; 101: 1520–1529.
Gertz MA, Lacy MQ, Dispenzieri A, Greipp PR, Litzow MR, Henderson KJ et al. Clinical implications of t(11;14)(q13;q32), t(4;14)(p16 3 q32) and -17p13 in myeloma patients treated with high-dose therapy. Blood 2005; 106: 2837–2840.
Jagannath S, Richardson PG, Sonneveld P, Schuster MW, Irwin D, Stadtmauer EA et al. Bortezomib appears to overcome the poor prognosis conferred by chromosome 13 deletion in phase 2 and 3 trials. Leukemia 2007; 21: 151–157.
San Miguel JF, Schlag R, Khuageva NK, Dimopoulos MA, Shpilberg O, Kropff M et al. Bortezomib plus melphalan and prednisone for initial treatment of multiple myeloma. N Engl J Med 2008; 359: 906–917.
Chng WJ, Kuehl WM, Bergsagel PL, Fonseca R . Translocation t(4;14) retains prognostic significance even in the setting of high-risk molecular signature. [published erratum appears in Leukemia 2008; 22: 462] Leukemia 2008; 22: 459–461, e-pub ahead of print 6 September 2007. No abstract available.
Trudel S, Li ZH, Wei E, Wiesmann M, Chang H, Chen C et al. CHIR-258, a novel, multitargeted tyrosine kinase inhibitor for the potential treatment of t(4;14) multiple myeloma. Blood 2005; 105: 2941–2948.
Shi J, Tricot GJ, Garg TK, Malaviarachchi PA, Szmania SM, Kellum RE et al. Bortezomib down-regulates the cell-surface expression of HLA class I and enhances natural killer cell-mediated lysis of myeloma. Blood 2008; 111: 1309–1317.
Keats JJ, Fonseca R, Chesi M, Schop R, Baker A, Chng WJ et al. Promiscuous mutations activate the noncanonical NF-kappaB pathway in multiple myeloma. Cancer Cell 2007; 12: 131–144.
Kuehl WM, Bergsagel PL . Multiple myeloma: evolving genetic events and host interactions. Nature Rev Cancer 2002; 2: 175–187.
Fonseca R, Debes-Marun CS, Picken EB, Dewald GW, Bryant SC, Winkler JM et al. The recurrent IgH translocations are highly associated with nonhyperdiploid variant multiple myeloma. Blood 2003; 102: 2562–2567.
Chng WJ, Winkler JM, Greipp PR, Jalal SM, Bergsagel PL, Chesi M et al. Ploidy status rarely changes in myeloma patients at disease progression. Leuk Res 2006; 30: 266–271.
Chng WJ, Van Wier SA, Ahmann GJ, Winkler JM, Jalal SM, Bergsagel PL et al. A validated FISH trisomy index demonstrates the hyperdiploid and nonhyperdiploid dichotomy in MGUS. Blood 2005; 106: 2156–2161.
Brousseau M, Leleu X, Gerard J, Gastinne T, Godon A, Genevieve F et al. Hyperdiploidy is a common finding in monoclonal gammopathy of undetermined significance and monosomy 13 is restricted to these hyperdiploid patients. Clin Cancer Res 2007; 13: 6026–6031.
Li X, Pennisi A, Zhan F, Sawyer JR, Shaughnessy JD, Yaccoby S . Establishment and exploitation of hyperdiploid and non-hyperdiploid human myeloma cell lines. Br J Haematol 2007; 138: 802–811.
Avet-Loiseau H, Li C, Magrangeas F, Gouraud W, Charbonnel C, Harousseau J-L et al. J Clin Oncol. Published online 17 August 2009, doi:10.1200/JCO.2008.20.6136.
Chng WJ, Kumar S, Vanwier S, Ahmann G, Price-Troska T, Henderson K et al. Molecular dissection of hyperdiploid multiple myeloma by gene expression profiling. Cancer Res 2007; 67: 2982–2989.
Hoyer JD, Hanson CA, Fonseca R, Greipp PR, Dewald GW, Kurtin PJ . The (11;14)(q13;q32) translocation in multiple myeloma A morphologic and immunohistochemical study. Am J Clin Pathol 2000; 113: 831–837.
Garand R, Avet-Loiseau H, Accard F, Moreau P, Harousseau J, Bataille R . t(11;14) and t(4;14) translocations correlated with mature lymphoplasmocytoid and immature morphology, respectively, in multiple myeloma. Leukemia 2003; 17: 2032–2035.
Fonseca R, Harrington D, Oken M, Kyle R, Dewald G, Bailey R et al. Myeloma and the t(11;14)(q13;q32) represents a uniquely defined biological subset of patients. Blood 2002; 99: 3735–3741.
Sawyer JR, Lukacs JL, Thomas EL, Swanson CM, Goosen LS, Sammartino G et al. Multicolour spectral karyotyping identifies new translocations and a recurring pathway for chromosome loss in multiple myeloma. Br J Haematol 2001; 112: 167–174.
Chesi M, Nardini E, Brents LA, Schrock E, Ried T, Kuehl WM et al. Frequent translocation t(4;14)(p16 3 q32 3 ) in multiple myeloma is associated with increased expression and activating mutations of fibroblast growth factor receptor 3. Nat Genet 1997; 16: 260–264.
Chesi M, Nardini E, Lim R, Smith K, Kuehl W, Bergsagel P . The t(4;14) translocation in myeloma dysregulates both FGFR3 and a novel gene, MMSET, resulting in IgH/MMSET hybrid transcripts. Blood 1998; 92: 3025–3034.
Chang H, Sloan S, Li D, Zhuang L, Yi QL, Chen CI et al. The t(4;14) is associated with poor prognosis in myeloma patients undergoing autologous stem cell transplant. Br J Haematol 2004; 125: 64–68.
Chang H, Qi XY, Samiee S, Yi QL, Chen C, Trudel S et al. Genetic risk identifies multiple myeloma patients who do not benefit from autologous stem cell transplantation. Bone Marrow Transplant 2005; 36: 793–796.
Trudel S, Ely S, Farooqi Y, Affer M, Robbiani DF, Chesi M et al. Inhibition of fibroblast growth factor receptor 3 induces differentiation and apoptosis in t(4;14) myeloma. Blood 2004; 103: 3521–3528.
Avet-Loiseau H, Facon T, Grosbois B, Magrangeas F, Rapp MJ, Harousseau JL et al. Oncogenesis of multiple myeloma: 14q32 and 13q chromosomal abnormalities are not randomly distributed, but correlate with natural history, immunological features, and clinical presentation. Blood 2002; 99: 2185–2191.
Fonseca R, Oken MM, Greipp PR . The t(4;14)(p16.3;q32) is strongly associated with chromosome 13 abnormalities in both multiple myeloma and monoclonal gammopathy of undetermined significance. Blood 2001; 98: 1271–1272.
Avet-Loiseau H, Facon T, Daviet A, Godon C, Rapp MJ, Harousseau JL et al. 14q32 translocations and monosomy 13 observed in monoclonal gammopathy of undetermined significance delineate a multistep process for the oncogenesis of multiple myeloma. Intergroupe Francophone du Myelome. Cancer Res 1999; 59: 4546–4550.
Fonseca R, Bailey RJ, Ahmann GJ, Rajkumar SV, Hoyer JD, Lust JA et al. Genomic abnormalities in monoclonal gammopathy of undetermined significance.[see comment]. Blood 2002; 100: 1417–1424.
Tricot G, Barlogie B, Jagannath S, Bracy D, Mattox S, Vesole DH et al. Poor prognosis in multiple myeloma is associated only with partial or complete deletions of chromosome 13 or abnormalities involving 11q and not with other karyotype abnormalities. Blood 1995; 86: 4250–4256.
Tricot G, Sawyer JR, Jagannath S, Desikan KR, Siegel D, Naucke S et al. Unique role of cytogenetics in the prognosis of patients with myeloma receiving high-dose therapy and autotransplants. J Clin Oncol 1997; 15: 2659–2666.
Perez-Simon JA, Garcia-Sanz R, Tabernero MD, Almeida J, Gonzalez M, Fernandez-Calvo J et al. Prognostic value of numerical chromosome aberrations in multiple myeloma: A FISH analysis of 15 different chromosomes. Blood 1998; 91: 3366–3371.
Zojer N, Konigsberg R, Ackermann J, Fritz E, Dallinger S, Kromer E et al. Deletion of 13q14 remains an independent adverse prognostic variable in multiple myeloma despite its frequent detection by interphase fluorescence in situ hybridization. Blood 2000; 95: 1925–1930.
Dewald GW, Therneau T, Larson D, Lee YK, Fink S, Smoley S et al. Relationship of patient survival and chromosome anomalies detected in metaphase and/or interphase cells at diagnosis of myeloma. Blood 2005; 106: 3553–3558.
Fonseca R, Oken MM, Harrington D, Bailey RJ, Van Wier SA, Henderson KJ et al. Deletions of chromosome 13 in multiple myeloma identified by interphase FISH usually denote large deletions of the q arm or monosomy. Leukemia 2001; 15: 981–986.
Fonseca R, Harrington D, Oken M, Dewald G, Bailey R, Van Wier S et al. Biologic and prognostic significance of interphase FISH detection of chromosome 13 abnormalities (D13) in multiple myeloma: an Eastern Cooperative Oncology Group (ECOG) Study. Cancer Res 2002; 62: 715–720.
Avet-Loiseau H, Li JY, Morineau N, Facon T, Brigaudeau C, Harousseau JL et al. Monosomy 13 is associated with the transition of monoclonal gammopathy of undetermined significance to multiple myeloma. Intergroupe Francophone du Myelome. Blood 1999; 94: 2583–2589.
Avet-Loiseau H, Daviet A, Saunier S, Bataille R . Chromosome 13 abnormalities in multiple myeloma are mostly monosomy 13. Br J Haematol 2000; 111: 1116–1117.
Gutierrez NC, Castellanos MV, Martin ML, Mateos MV, Hernandez JM, Fernandez M et al. Prognostic and biological implications of genetic abnormalities in multiple myeloma undergoing autologous stem cell transplantation: t(4;14) is the most relevant adverse prognostic factor, whereas RB deletion as a unique abnormality is not associated with adverse prognosis. Leukemia 2007; 21: 143–150.
Chiecchio L, Protheroe RK, Ibrahim AH, Cheung KL, Rudduck C, Dagrada GP et al. Deletion of chromosome 13 detected by conventional cytogenetics is a critical prognostic factor in myeloma. Leukemia 2006; 20: 1610–1617.
Chng WJ, Santana-Davila R, Van Wier SA, Ahmann GJ, Jalal SM, Bergsagel PL et al. Prognostic factors for hyperdiploid-myeloma: effects of chromosome 13 deletions and IgH translocations. Leukemia 2006; 20: 807–813.
Drach J, Ackermann J, Fritz E, Kromer E, Schuster R, Gisslinger H et al. Presence of a p53 gene deletion in patients with multiple myeloma predicts for short survival after conventional-dose chemotherapy. Blood 1998; 92: 802–809.
Chang H, Qi C, Yi QL, Reece D, Stewart AK . p53 gene deletion detected by fluorescence in situ hybridization is an adverse prognostic factor for patients with multiple myeloma following autologous stem cell transplantation. Blood 2005; 105: 358–360.
Chang H, Sloan S, Li D, Keith Stewart A . Multiple myeloma involving central nervous system: high frequency of chromosome 17p13.1 (p53) deletions. Br J Haematol 2004; 127: 280–284.
Tiedemann RE, Gonzalez-Paz N, Kyle RA, Santana-Davila R, Price-Troska T, Van Wier SA et al. Genetic aberrations and survival in plasma cell leukemia. Leukemia 2008; 22: 1044–1052.
Xiong W, Wu X, Starnes S, Johnson SK, Haessler J, Wang S et al. An analysis of the clinical and biologic significance of TP 53 loss and the identification of potential novel transcriptional targets of TP53 in multiple myeloma. Blood 2008; 112: 4235–4246.
Schilling G, Hansen T, Shimoni A, Zabelina T, Perez-Simon JA, Gutierrez NC et al. Impact of genetic abnormalities on survival after allogeneic hematopoietic stem cell transplantation in multiple myeloma. Leukemia 2008; 22: 1250–1255.
Shaughnessy Jr JD, Zhan F, Burington BE, Huang Y, Colla S, Hanamura I . A validated gene expression model of high-risk multiple myeloma is defined by deregulated expression of genes mapping to chromosome 1. Blood 2007; 109: 2276–2284.
Sawyer JR, Tricot G, Mattox S, Jagannath S, Barlogie B . Jumping translocations of chromosome 1q in multiple myeloma: evidence for a mechanism involving decondensation of pericentromeric heterochromatin. Blood 1998; 91: 1732–1741.
Hanamura I, Stewart JP, Huang Y, Zhan F, Santra M, Sawyer JR et al. Frequent gain of chromosome band 1q21 in plasma-cell dyscrasias detected by fluorescence in situ hybridization: incidence increases from MGUS to relapsed myeloma and is related to prognosis and disease progression following tandem stem-cell transplantation. Blood 2006; 108: 1724–1732.
Fonseca R, Van Wier SA, Chng WJ, Ketterling R, Lacy MQ, Dispenzieri A et al. Prognostic value of chromosome 1q21 gain by fluorescent in situ hybridization and increase CKS1B expression in myeloma. Leukemia 2006; 20: 2034–2040.
Annunziata CM, Davis RE, Demchenko Y, Bellamy W, Gabrea A, Zhan F et al. Frequent engagement of the classical and alternative NF-kappaB pathways by diverse genetic abnormalities in multiple myeloma. Cancer Cell 2007; 12: 115–130.
Liu P, Leong T, Quam L, Billadeau D, Kay NE, Greipp P et al. Activating mutations of N- and K-ras in multiple myeloma show different clinical associations: analysis of the Eastern Cooperative Oncology Group Phase III Trial. Blood 1996; 88: 2699–2706.
Bezieau S, Devilder MC, Avet-Loiseau H, Mellerin MP, Puthier D, Pennarun E et al. High incidence of N and K-Ras activating mutations in multiple myeloma and primary plasma cell leukemia at diagnosis. Human Mutat 2001; 18: 212–224.
Rasmussen T, Kuehl M, Lodahl M, Johnsen HE, Dahl IM . Possible roles for activating RAS mutations in the MGUS to MM transition and in the intramedullary to extramedullary transition some plasma cell tumors. Blood 2005; 105: 317–323.
Chng WJ, Gonzalez-Paz N, Price-Troska T, Jacobus S, Rajkumar SV, Oken MM et al. Clinical and biological significance of RAS mutations in multiple myeloma. Leukemia 2008; 22: 2280–2284.
Urashima M, Teoh G, Ogata A, Chauhan D, Treon SP, Sugimoto Y et al. Characterization of p16(INK4A) expression in multiple myeloma and plasma cell leukemia. Clin Cancer Res 1997; 3: 2173–2179.
Tasaka T, Asou H, Munker R, Said JW, Berenson J, Vescio RA et al. Methylation of the p16INK4A gene in multiple myeloma. Br J Haematol 1998; 101: 558–564.
Mateos MV, Garcia-Sanz R, Lopez-Perez R, Moro MJ, Ocio E, Hernandez J et al. Methylation is an inactivating mechanism of the p16 gene in multiple myeloma associated with high plasma cell proliferation and short survival. Br J Haematol 2002; 118: 1034–1040.
Chen W, Wu Y, Zhu J, Liu J, Tan S, Xia C . Methylation of p16 and p15 genes in multiple myeloma. Chi Med Sci J 2002; 17: 101–105.
Guillerm G, Depil S, Wolowiec D, Quesnel B . Different prognostic values of p15(INK4b) and p16(INK4a) gene methylations in multiple myeloma. Haematologica 2003; 88: 476–478.
Uchida T, Kinoshita T, Ohno T, Ohashi H, Nagai H, Saito H . Hypermethylation of p16 INK4A gene promoter during the progression of plasma cell dyscrasia. Leukemia 2001; 15: 157–165.
Gonzalez-Paz N, Chng WJ, McClure RF, Blood E, Oken MM, Van Ness B et al. Tumor suppressor p16 methylation in multiple myeloma: biological and clinical implications. Blood 2007; 109: 1228–1232.
Sarasquete ME, Garcia-Sanz R, Armellini A, Fuertes M, Martin-Jimenez P, Sierra M et al. The association of increased p14ARF/p16INK4a and p15INK4a gene expression with proliferative activity and the clinical course of multiple myeloma. Haematologica 2006; 91: 1551–1554.
Pichiorri F, Suh SS, Ladetto M, Kuehl M, Palumbo T, Drandi D et al. MicroRNAs regulate critical genes associated with multiple myeloma pathogenesis. Proc Natl Acad Sci USA 2008; 105: 12885–12890.
Loffler D, Brocke-Heidrich K, Pfeifer G, Stocsits C, Hackermuller J, Kretzschmar AK et al. Interleukin-6 dependent survival of multiple myeloma cells involves the Stat3-mediated induction of microRNA-21 through a highly conserved enhancer. Blood 2007; 110: 1330–1333.
Walker BA, Leone PE, Jenner MW, Li C, Gonzalez D, Johnson DC et al. Integration of global SNP-based mapping and expression arrays reveals key regions, mechanisms, and genes important in the pathogenesis of multiple myeloma. Blood 2006; 108: 1733–1743.
Jenner MW, Leone PE, Walker BA, Ross FM, Johnson DC, Gonzalez D et al. Gene mapping and expression analysis of 16q loss of heterozygosity identifies WWOX and CYLD as being important in determining clinical outcome in multiple myeloma. Blood 2007; 110: 3291–3300.
Mateos MV, Hernandez JM, Hernandez MT, Gutierrez NC, Palomera L, Fuertes M et al. Bortezomib plus melphalan and prednisone in elderly untreated patients with multiple myeloma: results of a multicenter phase I/II study. Blood 2006; 108: 2165–2172.
Chng WJ, Ahmann GJ, Henderson K, Santana-Davila R, Greipp PR, Gertz MA et al. Clinical implication of centrosome amplification in plasma cell neoplasm. Blood 2006; 107: 3669–3675.
Ahmann GJ, Jalal SM, Juneau AL, Christensen ER, Hanson CA, Dewald GW et al. A novel three-color, clone-specific fluorescence in situ hybridization procedure for monoclonal gammopathies. Cancer Genet Cytogenet 1998; 101: 7–11.
Harrison CJ, Mazzullo H, Ross FM, Cheung KL, Gerrard G, Harewood L et al. Translocations of 14q32 and deletions of 13q14 are common chromosomal abnormalities in systemic amyloidosis. Br J Haematol 2002; 117: 427–435.
Hayman SR, Bailey RJ, Jalal SM, Ahmann GJ, Dispenzieri A, Gertz MA et al. Translocations involving heavy-chain locus are possible early genetic events in patients with primary systemic amyloidosis. Blood 2001; 98: 2266–2268.
Avet-Loiseau H, Garand R, Lode L, Harousseau J-L, Bataille R . Translocation t(11;14)(q13;q32) is the hallmark of IgM, IgE, and nonsecretory multiple myeloma variants. Blood 2003; 101: 1570–1571.
Schop RF, Van Wier SA, Xu R, Ghobrial I, Ahmann GJ, Greipp PR et al. 6q deletion discriminates Waldenstrom macroglobulinemia from IgM monoclonal gammopathy of undetermined significance. Cancer Genet Cytogenet 2006; 169: 150–153.
Chng WJ, Glebov O, Bergsagel PL, Kuehl WL . Genetic events in the pathogenesis of multiple myeloma. Best Prac Res Clin Haematol 2007; 20: 571–596.
Author information
Authors and Affiliations
Corresponding author
Additional information
Members of the IMWG are listed in the appendix.
Appendix
Appendix
Rafat Abonour, Indiana University School of Medicine, Indianapolis, IN, USA
Ray Alexanian, MD Anderson, Houston, TX, USA
Kenneth Anderson, DFCI, Boston, MA, USA
Michael Attal, Purpan Hospital, Toulouse, France
Herve Avet-Loiseau, Institute de Biologie, Nantes, France
Ashraf Badros, University of Maryland, Baltimore, MD, USA
Leif Bergsagel, Mayo Clinic Scottsdale, Scottsdale, AZ, USA
Joan Bladé, Hospital Clinica, Barcelona, Spain
Bart Barlogie, MIRT UAMS Little Rock, AR, USA
Regis Batille, Institute de Biologie, Nantes, France
Meral Beksac, Ankara University, Ankara, Turkey
Andrew Belch, Cross Cancer Institute, Alberta, Canada
Bill Bensinger, Fred Hutchinson Cancer Center, Seattle, WA, USA
Mario Boccadoro, University of Torino, Torino, Italy
Michele Cavo, Universita di Bologna, Bologna, Italy
Wen Ming Chen, MM Research Center of Beijing, Beijing, China
Tony Child, Leeds General Hospital, Leeds, UK
James Chim, Department of Medicine, Queen Mary Hospital, Hong Kong
Ray Comenzo, Tufts Medical Center, Boston, MA, USA
John Crowley, Cancer Research and Biostatistics, Seattle, WA, USA
William Dalton, H Lee Moffitt, Tampa, FL, USA
Faith Davies, Royal Marsden Hospital, London, England
Cármino de Souza, Univeridade de Campinas, Caminas, Brazil
Michel Delforge, University Hospital Gasthuisberg, Leuven, Belgium
Meletios Dimopoulos, Alexandra Hospital, Athens, Greece
Angela Dispenzieri, Mayo Clinic, Rochester, MN, USA
Brian GM Durie, Cedars-Sinai Outpatient Cancer Center, Los Angeles, CA, USA
Hermann Einsele, Universitätsklinik Würzburg, Würzburg, Germany
Theirry Facon, Centre Hospitalier Regional Universitaire de Lille, Lille, France
Dorotea Fantl, Socieded Argentinade Hematolgia, Buenos Aires, Argentina
Jean-Paul Fermand, Hopitaux de Paris, Paris, France
Rafael Fonseca, Mayo Clinic Arizona, Scottsdale, AZ, USA
Gosta Gahrton, Karolinska Institute for Medicine, Huddinge, Sweden
Morie Gertz, Mayo Clinic, Rochester, MN, USA
John Gibson, Royal Prince Alfred Hospital, Sydney, Australia
Sergio Giralt, MD Anderson Cancer Center, Houston, TX, USA
Hartmut Goldschmidt, University Hospital Heidelberg, Heidelberg, Germany
Philip Greipp, Mayo Clinic, Rochester, MN, USA
Roman Hajek, Brno University, Brno, Czech Republic
Izhar Hardan, Tel Aviv University, Tel Aviv, Israel
Jean-Luc Harousseau, Institute de Biologie, Nantes, France
Hiroyuki Hata, Kumamoto University Hospital, Kumamoto, Japan
Yutaka Hattori, Keio University School of Medicine, Tokyo, Japan
Joy Ho, Royal Prince Alfred Hospital, Sydney, Australia
Vania Hungria, Clinica San Germano, Sao Paolo, Brazil
Shinsuke Ida, Nagoya City University Medical School, Nagoya, Japan
Peter Jacobs, Constantiaberg Medi-Clinic, Plumstead, South Africa
Sundar Jagannath, St Vincent's Comprehensive Cancer Center, New York, NY, USA
Hou Jian, Shanghai Chang Zheng Hospital, Shanghai, China
Douglas Joshua, Royal Prince Alfred Hospital, Sydney, Australia
Michio Kawano, Yamaguchi University, Ube, Japan
Nicolaus Kröger, University Hospital Hamburg, Hamburg, Germany
Shaji Kumar, Department of Hematology, Mayo Clinic, MN, USA
Robert Kyle, Department of Laboratory Med. and Pathology, Mayo Clinic, MN, USA
Juan Lahuerta, Grupo Espanol di Mieloma, Hospital Universitario, Madrid, Spain
Jae Hoon Lee, Gachon University Gil Hospital, Incheon, Korea
Xavier LeLeu, Hospital Huriez, CHRU Lille, France
Suzanne Lentzsch, University of Pittsburgh, Pittsburgh, PA, USA
Henk Lokhorst, University Medical CenterUtrecht, Utrecht, The Netherlands
Sagar Lonial, Emory University Medical School, Atlanta, GA, USA
Heinz Ludwig, Wilhelminenspital Der Stat Wien, Vienna, Austria
Angelo Maiolino, Rua fonte da Saudade, Rio de Janeiro, Brazil
Maria Mateos, University of Salamanca, Salamanca, Spain
Jayesh Mehta, Northwestern University, Chicago, IL, USA
GianPaolo Merlini, University of Pavia, Pavia, Italy
Joseph Mikhael, Mayo Clinic Arizona, Scottsdale, AZ, USA
Philippe Moreau, University Hospital, Nantes, France
Gareth Morgan, Royal Marsden Hospital, London, England
Nikhil Munshi, Diane Farber Cancer Institute, Boston, MA, USA
Ruben Niesvizky, Weill Medical College of Cornell University, New York, NY, USA
Yana Novis, Hospital SírioLibanês, Bela Vista, Brazil
Amara Nouel, Hospital Rutz y Paez, Bolivar, Venezuela
Robert Orlowski, MD Anderson Cancer Center, Houston, TX, USA
Antonio Palumbo, Cathedra Ematologia, Torino, Italy
Santiago Pavlovsky, Fundaleu, Buenos Aires, Argentina
Linda Pilarski, University of Alberta, Alberta, Canada
Raymond Powles, Leukaemia & Myeloma, Wimbledon, England
S. Vincent Rajkumar, Mayo Clinic, Rochester, MN, USA
Donna Reece, Princess Margaret Hospital, Toronto, Canada
Tony Reiman, Cross Cancer Institute, Alberta, Canada
Paul Richardson, Dana Farber Cancer Institute, Boston, MA, USA
Angelina Rodriquez Morales, Bonco Metro Politano de Sangre, Caracas, Venezuela
Orhan Sezer, Department of Hem/Onc, Universitatsklinikum Charite, Berlin, Germany
John Shaughnessy, MIRT UAMS, Little Rock, AR, USA
Kazuyuki Shimizu, Nagoya City Midori General Hospital, Nagoya, Japan
David Siegel, Hackensack, Cancer Center, Hackensack, NJ, USA
Jesus San Miguel, University of Salamanca, Salamanca, Spain
Chaim Shustik, McGill University, Montreal, Canada
Seema Singhal, Northwestern University, Chicago, IL, USA
Pieter Sonneveld, Erasmus MC, Rotterdam, The Netherlands
Andrew Spencer, The Alfred Hospital, Melbourne, Australia
Edward Stadtmauer, University of Pennsylvania, Philadelphia, PA, USA
A Keith Stewart, Mayo Clinic Arizona, Scottsdale, AZ, USA
Patrizia Tosi, Italian Cooperative Group, Istituto di Ematologia Seragnoli, Bologna, Italy
Guido Tricot, Huntsman Cancer Institute, Salt Lake City, UT, USA
Ingemar Turesson, Department of Hematology, Malmo University, Malmo, Sweden
Brian Van Ness, University of Minnesota, Minneapolis, MN, USA
Ivan Van Riet, Brussels Vrija University, Brussels, Belgium
Robert Vescio, Cedars-Sinai Cancer Center, Los Angeles, CA, USA
David Vesole, Loyola University Chicago, IL, USA
Anders Waage, University Hospital, Trondheim, Norway NSMG
Michael Wang, MD Anderson, Houston, TX, USA
Donna Weber, MD Anderson, Houston, TX, USA
Jan Westin, Sahlgrenska University Hospital, Gothenburg, Sweden
Keith Wheatley, University of Birmingham, Birmingham, UK
Dina B Yehuda, Department of Hematology, Hadassah University Hospital, Hadassah, Israel
Jeffrey Zonder, SWOG, Department of Hem/Onc., Karmanos Cancer Institute, MI, USA
Rights and permissions
About this article
Cite this article
Fonseca, R., Bergsagel, P., Drach, J. et al. International Myeloma Working Group molecular classification of multiple myeloma: spotlight review. Leukemia 23, 2210–2221 (2009). https://doi.org/10.1038/leu.2009.174
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2009.174
Keywords
This article is cited by
-
Prognostic factors in 448 newly diagnosed multiple myeloma receiving bortezomib-based induction: impact of ASCT, transplant refusal and high-risk MM
Bone Marrow Transplantation (2024)
-
ML-based sequential analysis to assist selection between VMP and RD for newly diagnosed multiple myeloma
npj Precision Oncology (2023)
-
Imaging flow cytometry-based multiplex FISH for three IGH translocations in multiple myeloma
Journal of Human Genetics (2023)
-
Integrated analysis of next generation sequencing minimal residual disease (MRD) and PET scan in transplant eligible myeloma patients
Blood Cancer Journal (2023)
-
Daratumumab with lenalidomide and dexamethasone in relapsed or refractory multiple myeloma patients – real world evidence analysis
Annals of Hematology (2023)