Abstract
Alternative splicing of the human glucocorticoid receptor gene generates two isoforms, hGRα and hGRβ. hGRβ functions as a dominant-negative regulator of hGRα activity and but also has inherent transcriptional activity, collectively altering glucocorticoid sensitivity. Single-nucleotide polymorphisms in the 3′ UTR of hGRβ have been associated with altered receptor protein expression, glucocorticoid sensitivity, and disease risk. Characterization of the hGRβ G3134T polymorphism has been limited to a relatively small, homogenous population. The objective of this study was to determine the prevalence of hGRβ G3134T in a diverse population and assess the association of hGRβ G3134T in this population with physiological outcomes. In a prospective cohort study, 3730 genetically diverse participants were genotyped for hGRβ G3134T and four common GR polymorphisms. A subset of these participants was evaluated for clinical and biochemical measurements. Immortalized human osteosarcoma cells (U-2 OS), stably transfected with wild-type or G3134T hGRβ, were evaluated for receptor expression, stability, and genome-wide gene expression. Glucocorticoid-mediated gene expression profiles were investigated in primary macrophages isolated from participants. In a racially diverse population, the minor allele frequency was 74% (50.7% heterozygous carriers and 23.3% homozygous minor allele), with a higher prevalence in Caucasian non-Hispanic participants. After adjusting for confounding variable, carriers of hGRβ G3134T were more likely to self-report allergies, have higher serum cortisol levels, and reduced cortisol suppression in response to low-dose dexamethasone. The presence of hGRβ G3134T in U-2 OS cells increased hGR mRNA stability and protein expression. Microarray analysis revealed that the presence of the hGRβ G3134T polymorphism uniquely altered gene expression profiles in U-2 OS cells and primary macrophages. hGRβ G3134T is significantly present in the study population and associated with race, self-reported disease, and serum levels of glucocorticoids. Underlying these health differences may be changes in gene expression driven by altered receptor stability.
Similar content being viewed by others
Main
Glucocorticoids are primary mediators of physiological homeostasis, and, as such, regulate a wide array of processes throughout the body. The powerful actions of corticosteroids make them one of the most commonly prescribed classes of drugs worldwide. One of the primary homeostatic functions of glucocorticoids is regulating the immune system, in which glucocorticoids display potent pro- and anti-inflammatory actions.1 Precise regulation over these actions is necessary, as gross glucocorticoid imbalances can lead to pathological conditions, such as Cushing’s syndrome or Addison’s disease. In addition, subtle changes in glucocorticoid signaling have been implicated in many common immune-mediated diseases, including insulin resistance, hypertension, metabolic syndrome, depression, and autoimmune or allergic disorders.2 Moreover, individual variability in the response to glucocorticoids alters their therapeutic efficacy.
The physiological actions of glucocorticoids are mediated by the glucocorticoid receptor (GR; NR3C1), a ligand-dependent transcription factor that regulates global gene expression by direct DNA binding or through interactions with other transcription factors. The pleotropic actions ascribed to GR are a result of the cellular processing of the GR gene and protein. The human GR gene is subject to alternative splicing at the 3′ end, giving rise to two receptor isoforms (hGRα and hGRβ) that differ in their carboxyl terminal.3 Although hGRβ does not bind glucocorticoids, it has intrinsic transcriptional activity and functions as a dominant-negative inhibitor of the classic GR hGRα.4, 5, 6
Individual sensitivity to glucocorticoid signaling is also determined by genetic variability. Several studies have identified polymorphisms in the hGR gene that are associated with altered glucocorticoid function. The clinical features associated with these polymorphisms include increased inflammatory parameters, cardiovascular disease, rheumatoid arthritis, metabolic disorders, and psychiatric diseases7, 8, 9, 10 (reviewed in Koper et al11). The mechanisms by which hGR polymorphisms result in disease are not clear, although some studies have suggested changes to the intracellular ratio of hGRα and hGRβ.12 The A3669G hGRβ (rs6198) polymorphism in the 3′ UTR of exon 9β stabilizes hGRβ and is associated with immunologically related diseases.12, 13 An additional hGRβ polymorphism G3134T in the 3′ UTR (rs6191) has been identified. However, reports are limited to two case–control association studies in a relative homogenous population.8, 14 No studies have been performed on the frequency of hGRβ G3134T in a diverse population, and the functional role of this polymorphism is not known.
In this study, we assessed the distribution of five hGR polymorphisms, including hGRβ G3134T, in a racially diverse population. Self-reported health outcomes and serum laboratory values were evaluated based on the hGRβ G3134T genotype. To address the functional consequence of hGRβ G3134T we performed in vitro analysis on stably transfected human cells and peripheral blood mononuclear cells (PBMCs) isolated from participants. We hypothesized that hGRβ G3134T would be associated with immune-mediated outcomes and altered stability of the receptor. We report an association of the G3134T hGRβ variant with self-reported outcomes and serum cortisol levels, and a molecular basis for altered glucocorticoid sensitivity among individuals.
Materials and methods
Study Population
Studies were performed at the NIEHS Clinical Research Unit (CRU) and approved by the NIEHS Institutional Review Board (IRB 10-E-0130). DNA samples were obtained from the Environmental Polymorphisms Registry (EPR; IRB 04-E-0053), a large population cohort in North Carolina, and genotyped as described below.15 Participants included 3730 adults aged 18 years and older at the time of enrollment representing a racially and ethnically diverse population (stratified as Caucasian non-Hispanic, Caucasian Hispanic, Black African American, or Asian). In addition, the Environmental Polymorphism Registry Health and Exposure Survey was distributed to all EPR participants, and self-reported outcomes for 1781 of the 3730 above-mentioned participants were available for analysis. Additional clinical characteristics were obtained from a subgroup of the 3730 EPR participants. On-site screening procedures for this subgroup consisted of medical health history, physical examination, including height, weight, and blood tests. Inclusion criteria for the participants providing medical history and clinical samples were as follows: at least 18 years of age at the time of enrollment, healthy as defined by the International Red Cross guidelines, able to understand and provide informed consent, and willing and able to fast for up to 12 h. Females were excluded if they were pregnant or currently breast-feeding. Participants were also excluded if there was any glucocorticoid use within 14 days of the study visit. Participants were instructed to abstain from all food and drink, with the exception of water, for 12 h prior to the study visit (scheduled between 0800–0930 hours). Blood samples were processed, shipped, and stored according to the NIEHS CRU Laboratory Test Procedures and Standard Operating Procedures. C-reactive protein (CRP), total cholesterol, cholesterol fractionation (high-density lipoprotein: HDL and low-density lipoprotein: LDL), triglycerides, serum cortisol, serum creatinine, glucose, and insulin were measured at the CLIA-certified laboratory at the NIH Clinical Center (Bethesda, MD). Serum cortisol levels were measured from blood using a solid-phase competitive chemiluminescent enzyme immunoassay. Laboratory values obtained from 272 participants were analyzed based on genotype. After the first clinical visit, participants were considered for a modified dexamethasone suppression test (DST) at a second clinical visit based on the following exclusion criteria: currently enrolled in another clinical research study or receiving an investigational agent, use of medications containing corticosteroids, estrogens, or tetracyclines, uncontrolled hypertension, active coronary artery disease, renal failure, glaucoma, uncontrolled psychiatric disorders, active systemic fungal infection, ongoing cancer treatment, prior hypersensitivity to dexamethasone, or unwilling to take dexamethasone at scheduled time. Participants deemed eligible for the modified DST (138 total) were provided one tablet of 0.25 mg dexamethasone to take 9 h prior to their scheduled appointment. Participants were instructed to abstain from all food and drink, with the exception of water, for 12 h prior to the study visit (scheduled to occur at the documented time of the first clinical visit).
Genotyping
DNA was extracted from whole blood using the Gentra Autopure robot (Qiagen, Valencia, CA) as per the manufacturer’s protocol. DNA concentration was assessed using the DTX 880 plate reader (Beckman Coulter, Jersey City, NJ) and normalized to 50 ng/μl. Subsequent genotyping was performed by the NIEHS Molecular Genetics Core using the Illumina customized GoldenGate Assay (San Diego, CA) for the hGR single-nucleotide polymorphisms (SNP): ER22/23EK (rs6189/90), N363S (rs6195), Bcl1 (rs41423247), βG3134T (rs6191), and βA3669G (rs6198; Figure 1). Genotyping duplicate reproducibility was 99.76%. Genotyping was performed with the use of multiple controls to ensure quality and fidelity. Illumina internal controls including allele-specific extension, PCR uniformity, gender genotype, gap extension efficiency, annealing specificity, and hybridization were consistent with successful assay performance. PCR contamination controls were negative. In addition, samples were genotyped concurrently for gender and race markers to decrease the possibility of sample misidentification. Gender genotype mismatched with reported gender for 37 samples, and these samples were excluded from the study.
Reagents
Heat-inactivated fetal bovine serum (FBS) was purchased from Atlanta Biologicals (Lawrenceville, GA). Charcoal-stripped heat-inactivated FBS was purchased from Hyclone (Logan, UT). Dexamethasone (Dex; 1,4-Pregnadien-9-Fluoro-16-Methyl-11β,17,21-Triol-3,20-Dione) was purchased from Steraloids (Newport, RI). TaqMan RT-PCR primer-probes were purchased from Applied Biosystems (Foster City, CA).
Culture of Human U-2 0 S Cells Stably Expressing hGRα, hGRβ, or hGRβG3134T
The parental U-OFF cell line was established by transfecting the GR-null cell line U-2 OS (obtained from American Type Culture Collection, Manassas, VA) with the BD Clontech plasmid pTet-OFF plasmid (Clontech, Mountain View, CA). The generation of U-2 OS stably expressing hGRα has previously been described.16 To generate the WT hGRβ cells, MluI and EcoRV ends were added onto full length hGRβ by PCR amplification of the pCMVhGRβ plasmid.3 The pTRE2hyg vector was digested with MluI and EcoRV, and the two DNAs were ligated to form the pTRE2hGRβ plasmid. Site-directed mutagenesis was performed to make pTRE2hGRβG3134T. The wild-type (WT) pTRE2hGRβ and pTRE2hGRbG3134T plasmid were individually transfected into the U-OFF cells, and stably expressing clones were selected using 500 μg/ml of Geneticin and 500 μg/ml of hygromycin. Expression of the vector was compared in WT and G3134T hGRβ-expressing cells (Supplementary Figure 1). Expression of GR was compared for each of these cell lines using western blot analyses. U-2 OS stable cell lines, U-OFF, WT hGRβ, and G3134T hGRβ, were maintained in DMEM/F-12 supplemented with 10% FCS:CS, 2 mm glutamine, and 100 IU of penicillin and 100 mg/ml of streptomycin and grown in a standard tissue culture incubator at 37 °C, with 95% humidity and 5% carbon dioxide.17
Flow Cytometry
PBMCs were isolated from patient blood collected in a Vacuette K3 EDTA tube (Greiner Bio-One, Monroe, NC), using Histopaque-1077 (Sigma-Aldrich, St Louis, MO) density gradient centrifugation. Purified PBMCs were frozen at 5 × 106 cells per ml in cryopreservation media containing RPMI 1640, 11.25% human serum albumin (Gemini Bio-Products, West Sacramento, CA), and 10% DMSO (Sigma-Aldrich). Cells were stored in a Mr Frosty container (Thermo Fisher Scientific, Waltham, MA) at −80 °C for 24 h before transferring to liquid nitrogen storage. Frozen human PBMCs were thawed, washed twice with RPMI 1640 supplemented with 10% heat-inactivated FCS, 2 mM L-glutamine, 100 IU penicillin, and 100 μg/ml streptomycin, then rested overnight at 37 °C before staining. The following day, cells were treated with 2.5 μg Human Fc Block (BD Biosciences, San Jose, CA) for 20 min at room temperature. Samples were fixed and permeabilized for 20 min at 4 °C with 100 μl Cytofix/Cytoperm solution (BD Biosciences). Cells were incubated with anti-human GRβ receptor polyclonal antibody (0.04 mg/ml) for 20 min on ice, followed by a secondary goat anti-rabbit IgG APC (1:200; Thermo Fisher Scientific) for 20 min on ice. Thirty thousand cells were acquired on a BD FACS Aria II, standardized with Calibrite APC beads (BD Biosciences), and analyzed using FlowJo software (Treestar, Ashland, OR).
Immunoblotting
U-2 OS cells grown in vitro were lysed in RIPA buffer. Membranes with equivalent amounts of protein were incubated with polyclonal anti-GR antibodies (D8H2 1:1000; Cell Signaling, Danvers, MA) or monoclonal anti-β-actin antibodies (1:10 000; EMD Millipore, Billerica, MA). Quantification of immunoreactivity was determined by incubation with a mixture of goat anti-rabbit Alexa Fluorophore 680-conjugated (Molecular Probes, Carlsbad, CA) and goat anti-mouse IRDye 800-conjugated secondary (Rockland Immunochemicals, Gilbertsville, PA) antibodies for 1 h at room temperature and visualization using the Odyssey LiCor imaging system.
Immunofluorescence
Cells grown on glass-bottom plates were fixed, and nonspecific binding was blocked by incubation with 5% normal goat serum (Vector Laboratories, Burlingame, CA). Plates were incubated overnight with anti-GR antibodies (1:1000). The following day plates were washed and incubated with Alexa Fluor 488 anti-rabbit IgG (Molecular Probes) for 30 min. Plates underwent final washing, and nuclei were visualized by DRAQ5 (Cell Signaling) staining prior to visualization on the Zeiss LSM 780 (Carl Zeiss Microscopy, Jena, Germany) confocal microscope.
RNA Isolation and Quantitative Real-Time PCR
Total RNA was extracted from U-2 OS cells using the QIAGEN RNeasy Mini Kit (Qiagen) with deoxyribonuclease (DNase) treatment performed on-column using a Ribonuclease-Free DNase Kit (Qiagen), according to the manufacturer’s instruction. The abundance of mRNAs was determined from at least four biological replicates using predesigned real-time probes and primers (Applied Biosystems) on a 7900HT sequence detection system (Applied Biosystems). The signal obtained from each gene primer–probe set was normalized to that of the unregulated housekeeping gene peptidylprolyl isomerase B (PPIB) primer–probe set.
Total RNA was extracted from macrophages obtained from participants and treated for 6 h with vehicle or 100 nM dexamethasone using the QIAGEN RNeasy Mini Kit (Qiagen) with DNase treatment performed on-column using a Ribonuclease-Free DNase Kit (Qiagen), according to the manufacturer’s instruction. Macrophages were differentiated from mononuclear cells ex vivo as described previously.18 Briefly, whole blood was collected from study participants, and PBMCs were rapidly isolated using Histopaque-1077 according to the manufacturer’s protocol (Sigma-Aldrich). To differentiate mononuclear cells to macrophages, PBMCs were plated in X-VIVO medium (Lonza, Walkersville, MD) supplemented with 10% human serum, 100 IU of of penicillin, and 100 mg/ml of streptomycin and grown in a standard tissue culture incubator for 7 days. Medium was replaced every 3–4 days, and cells were treated on day 8. RNA was isolated from seven control participants and three participants homozygous for the minor allele of the G3134T hGRβ polymorphism.
Microarray Analysis
Gene expression analysis was conducted using Agilent Whole Human Genome 4x44 multiplex format oligo arrays (014850; Agilent Technologies) following the Agilent 1-color microarray-based gene expression analysis protocol. Starting with 500 ng of total RNA, Cy3-labeled cRNA was produced according to the manufacturer’s protocol. For each sample, 1.65 μg of Cy3-labeled cRNAs were fragmented and hybridized for 17 h in a rotating hybridization oven. Slides were washed and then scanned with an Agilent Scanner. Data were obtained using the Agilent Feature Extraction software (v9.5), using the 1-color defaults for all parameters. The Agilent Feature Extraction Software performed error modeling, adjusting for additive and multiplicative noise. Gene expression was analyzed with the Partek Genomics Suite (Partek, St Louis, MO, USA), to identify significantly regulated probes. Significance was indicated as any change relative to U-OFF cells with a P-value and false discovery rate (step up)<0.05 in U-2 OS cells. In the differentiated macrophages, significantly regulated genes were determined as any change in the samples from participants homozygous for the minor allele of the G3134T hGRβ polymorphism relative to vehicle-treated samples from participants genotyped as homozygous WT for the G3134T hGRβ polymorphism (control). Heat map analysis of significantly regulated probes was generated with the Partek Genomics Suite. Dendrograms of samples (columns) and genes (row) were generated by Hierarchical clustering. The lists of significant probe sets by genotype were visually sorted by Venn diagram (http://www.pangloss.com/seidel/Protocols/venn.cgi) and further analyzed by Ingenuity Pathway Analysis (IPA; version 6.5; Ingenuity Systems, Redwood City, CA). Gene enrichment P-values (P<0.05) for biological functions were determined by IPA using Fisher’s exact test. Significantly regulated genes were also analyzed for function by GO using GATHER.
Statistical Analysis
For each SNP, the number of patients with 0, 1, and 2 copies of the minor allele were reported for the overall cohort. A Pearson’s χ2-test was used to determine the association between hGRβ G3134T and race. The relationship between hGRβ G3134T and self-reported outcomes was examined by fitting a logistic regression model for the presence of a characteristic as a function of genotype. Specifically, the genotype was modeled as binary indicator for carriers vs non-carriers of the mutant allele, and characteristics were reported as binary endpoints when patients reported 'Yes' to one of the evaluated outcomes. For cardiovascular disease, several related self-reported outcomes were combined including: angina (28), heart attack (54), coronary artery disease (43), congestive heart failure (28), mini stroke (50), and stroke (25) for a total of 137 cardiovascular disease 'Yes' responses. Diabetes included participants who responded ‘Yes’ to both pre-diabetes and diabetes for a total of 323 'Yes' responses. The total 'Yes' responses for outcomes were as follows: allergies (yes=686), COPD (yes=66), asthma (yes=255), inflammatory bowel disease (Crohns/ulcerative colitis yes=40), autoimmune disease (fibromyalgia/lupus/Sjogrens yes=193), rheumatoid arthritis (yes=102), endometriosis (yes=105). Both unadjusted and adjusted models were fit. Adjusted models accounted for the following potential confounders: age, gender, race, height, and weight. The relationship between hGRβ G3134T and laboratory values was examined by fitting a linear regression model for laboratory finding as a function of the genotype (i.e., ANOVA). Specifically, the genotype was modeled as binary indicator for carriers vs non-carriers of the mutant allele. CRP was natural log-transformed to account for non-normal distribution of value. Both unadjusted and adjusted models were fit. Adjusted models accounted for the following potential confounders: sex, age, race, height, weight, and body mass index. All analyses were performed using Statistical Analysis Software (version 9.2; SAS Institute, Cary, NC). Gene expression data are presented as means±s.e.m. Statistical significance was determined by ANOVA with Tukey’s post hoc analysis. Statistical significance was defined as *P<0.05 or **P<0.01.
Results
Distribution of the GR Polymorphisms in a Racially Diverse Population
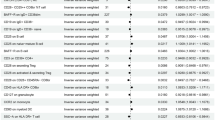
Table 1 shows EPR participant characteristics by age, gender, and race. The mean age of the EPR participants was 43.9 (±14.8) years. There were 1672 (44.8%) men and 2058 (55.2%) women. In our population of participants, 1829 (49%) were Caucasian non-Hispanic, 331 (8.9%) were Caucasian Hispanic, 1559 (41.8%) were Black African American (Black AA), and 11 (3%) reported their race as other. The overall genotyping rate for the six hGR SNPs was 99.08%. The most frequently observed polymorphism in the study population was rs6191 (G3134T hGRβ) with a frequency of 26% for the GG genotype, 50.7% for the GT genotype, and 23.3% for the TT genotype (overall minor allele frequency of 74%; Table 1). The frequency of the G3134T hGRβ polymorphism was not correlated with gender, but demonstrated a significant association with all races included. There was a greater percentage of Caucasian non-Hispanics with the minor T allele (TT>GT> GG). Conversely, the frequency of the T allele was less in the Caucasian Hispanic and Black AA participants (GG>GT>TT). These data indicate that incidence of the highly prevalent G3134T hGRβ polymorphism is associated with race.
Correlation of Genotype to Self-Reported Outcomes
Several polymorphisms of hGR are functionally relevant relating to altered glucocorticoid sensitivity and associated with inflammatory-related diseases.11 To determine the impact of the G3134T hGRβ polymorphism on clinical features, demographics and self-reported outcomes related to inflammation were evaluated from participants completing the Health and Exposure Survey (Table 2). Data were available from 1781 EPR participants. The prevalence of the G3134T hGRβ polymorphism was distributed at a 1:2:1 ratio in both genders. Weight and height were not significantly associated with the polymorphism. As with the entire genotyped cohort, race was significantly associated with allele frequency. Table 2 shows the number of respondents reporting a health outcome according to genotype. In an unadjusted model, there were no significant differences between genotypes for any reported outcome. After adjusting for potentially confounding variables, the G3134T hGRβ polymorphism was significantly associated with allergic rhinitis/allergies and approached significance for autoimmune disease (P=0.06).
Comparison of Clinical Parameters Among Carriers and Non-Carriers
Studies have reported associations between polymorphisms in the GR gene and laboratory markers of disease. Table 3 shows the laboratory values from participants with stored serum samples. The frequency of genotypes was equally distributed among both genders. Across genotypes, participants had similar measurement for height, weight, and waist circumference. In this smaller cohort, race remained significantly associated with genotype. No significant associations were observed between genotype and laboratory values of systolic or diastolic blood pressure, CRP, cholesterol, HDL, LDL, triglycerides, creatinine, glucose, and insulin. Significantly higher serum cortisol levels were found in participants heterozygous and homozygous for the minor allele (P=0.03). Statistical significance persisted after adjustment for confounding variables (P=0.02). Interestingly, assessment of adrenal function by the DST indicated that participants heterozygous and homozygous for the hGRβ G3134T polymorphism demonstrate reduced inhibition of cortisol compared to participants with homozygous for the major G3134T hGRβ allele (P<0.01 unadjusted and adjusted model).
In vitro Assessment of the hGRβ G3134T Polymorphism Reveals Altered mRNA Stability and Protein Expression
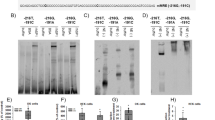
Polymorphisms in the 3′ UTR of hGRβ have been shown to alter mRNA stability, which is suggested to underlie the mechanism of associated disease.12 To determine whether the hGRβ G3134T polymorphism could modify hGRβ mRNA stability, hGRβ mRNA was measured in U-2 OS cells stably expressing either WT hGRβ or G3134T hGRβ that had been incubated with actinomycin D (Figure 2a). The stability of hGRβ mRNA was significantly increased in cells harboring the G3134T polymorphism (P<0.0001). The half-life of hGRβ mRNA in cells with the G3134T polymorphism was increased ~1.0 h compared to WT hGRβ (t1/2 WT hGRβ=72.42 min; t1/2 hGRβ G3134T=123.38 min). To evaluate whether the increase in mRNA stability resulted in increased steady-state hGRβ protein expression, hGRβ protein levels were measured by western blot (Figure 2b). Compared to the WT genotype, hGRβ protein levels were 33% higher, suggesting that increased mRNA stability resulting from the G3134T polymorphism results in significantly greater steady-state levels of hGRβ protein. To investigate whether increased expression of hGRβ established in vitro was also present in patients with two copies of the minor allele of the G3134T hGRβ polymorphism compared to patients homozygous for the major allele, levels of hGRβ were evaluated in human PBMCs by flow cytometry (Figure 2c). The numerical average of percent PBMC expressing hGRβ and median fluorescence intensity was higher in patients with two copies of the G3134T hGRβ polymorphism. The difference between the two genotypes was more apparent in the lymphocyte population of PBMCs compared to monocytes, which suggests that the consequences of harboring the G3134T hGRβ polymorphism may be cell-type-specific. Immunofluorescence and confocal microscopy were employed to determine whether increased levels of hGRβ or the G3134T polymorphism altered the subcellular distribution of hGRβ. Unlike hGRα, hGRβ resides primarily in the nucleus independent of hormone availability (Figure 2d).3 The G3134T polymorphism did not alter the subcellular distribution of hGRβ. To determine whether the presence of the G3134T polymorphism in hGRβ altered the ability of hGRα to translocate to the nucleus in response to ligand, U-2 OS cells stably expressing hGRα were transfected with WT hGRβ, G3134T hGRβ, or an empty vector control (Figure 2e). Nuclear translocation of hGR in cells expressing the G3134T hGRβ polymorphism following 1 h treatment with 100 nM dexamethasone was equivalent to cells transfected with empty vector control and cells expressing WT hGR β.
G3134T hGRβ polymorphism affects GR stability resulting in elevated GR protein expression. (a) Semi-log plot of relative mRNA expression vs time. RNA decay was measured by treating wild type (WT) hGRβ and G3134T hGRβ cells with 5 μg/μl of actinomycin D and harvesting mRNA at defined time points. Expression of hGRβ was measured by quantitative RT-PCR and values were normalized to the housekeeping gene Cyclophilin B (PPIB). mRNA expression for WT hGRβ at time 0 was set to 100% relative mRNA and expression at subsequent time points were set relative the expression at time 0. Data points show mean±s.e.m. of four biological replicates and **P<0.01. (b) hGRβ protein levels were quantified by western blot analysis and normalized to levels of the housekeeping protein β-actin (n=6–8 per genotype). The results represent the mean±s.e.m. and *P<0.05. (c) hGRβ levels were measured by flow cytometry and the distribution of data are displayed for the % of cells that stained positive for hGRβ and the median fluorescence intensity (MFI) for hGRβ-positive staining cells. Patients with no copies of rs6189, rs6190, rs41423247, rs6191, and rs6198 were used for the 0 copy group (n=10) and patients with two copies of rs6191 (G3134T hGRβ) and varying copy number of the other polymorphisms were used for the two copy group (n=9). Lymphocytes and monocytes were determined from the total Peripheral blood mononuclear cell (PBMC) population by forward and side scatter. (d) Immunofluorescence of the glucocorticoid receptor was performed in fixed U-2 OS cells transfected with WT or G3134T hGRβ. Glucocorticoid receptor expression is shown in green and DRAQ5 staining of the nuclei is shown in red. (e) Immunofluorescence of the glucocorticoid receptor (green) was performed in U-2 OS cells stably expressing hGRα transfected with empty vector, WT hGRβ, or G3134T hGRβ and treated for 1 h with 100 nM Dex. Images taken at × 630.
Immortalized Cells Harboring hGRβ G3134T Polymorphism Demonstrate altered Global Gene Expression
To assess whether the presence of the hGRβ G3134T polymorphism affected hGRβ signaling, genome-wide microarray analysis was performed on isolated mRNA from the U-OFF cells and U-2 OS cells stably expressing WT hGRβ and G3134T hGRβ. Visualization of variance by principal component analysis indicated that there was high concordance by genotype (Figure 3a). Furthermore, it was evident by the spatial distribution of samples that the hGRβ G3134T polymorphism significantly altered global gene expression. Gene lists were created in Partek Genomics Suite to identify significant differentially expressed genes. Surprisingly, only a third of the differentially expressed genes in the hGRβ G3134T cells were in common with the WT hGRβ cells (Figure 3a). Moreover, the single-nucleotide substitution in hGRβ resulted in 2649 uniquely expressed genes. Differences in the overall directionality of gene regulation were visualized by generating a heat map of significantly regulated genes in U-OFF, WT hGRβ, and G3134T hGRβ cells (red represents induced genes and green represents repressed genes). This revealed that the G3134T polymorphism had a minor effect on the ratio of induced and repressed genes (59.2% and 40.8%, respectively, compared to 62.6% and 37.4% of genes regulated in WT hGRβ cells).
Global gene expression is altered by G3134T hGRβ polymorphism. (a) mRNA isolated from three biological replicates of each genotype were analyzed by the Agilent Whole Human Genome 4 × 44 multiplex format oligo array for gene expression. Principal component analysis was performed in Partek Genomics Suite to visualize the variance within each genotype. The number of probes statistically different compared to U-OFF cells at P<0.05 with false discovery rate (step up) genotypes were sorted by Venn diagram. Heat map of significantly regulated gene probes by genotype was created with Partek Genomics Suite software and visually represent unique and commonly regulated targets. Red indicates induced genes and green indicates repressed genes. (b) All significantly regulated genes were analyzed by IPA software. A comparison analysis was performed, and the top 15 Diseases and Biological Functions and Canonical Signaling Pathways are reported. The activation score is a prediction of the activation state of the molecules in a given function. Orange bars predict an overall increase in the activity of the pathway, while blue bars indicate a prediction of an overall decrease in activity. (c) Genes significantly regulated in both genotypes (1319 total) were analyzed by IPA software. The top five Diseases and Biological Functions and Canonical Signaling Pathways are shown for each genotype. (d) A comparison analysis of genes uniquely regulated in either WT hGRβ cells or G3134T hGRβ cells was performed, and the top five Diseases and Biological Functions and Canonical Signaling Pathways are shown for each genotype. (e) Quantitative RT-PCR was performed on independent mRNA samples to validate expression changes in IL13RA2, CBLN2, TRAF1, and EDN3 mRNA from total isolated RNA. The reported fold change from microarray analysis is listed below the quantitative RT-PCR results. The results represent the mean±s.e.m. and *P<0.05/**P<0.01.
To define the biological pathways enriched by the transcripts associated with the G3134T polymorphism, literature-based IPA was performed on the gene lists from WT hGRβ (2078 genes) and G3134T hGRβ (4530 genes) cells. A comparison analysis revealed that the activation state of the top 15 Molecular and Cellular Functions was remarkably different in the presence of the G3134T polymorphism (Figure 3b). The top function in the comparison analysis was neonatal death, and it was predicted to have an activation z-score of 2.804 in G3134T hGRβ cells but an activation z-score of 0 in WT hGRβ cells. Visualizing the genes associated with this function revealed strong induction or repression of 99 genes in cells expressing the G3134T polymorphism (Supplementary Figure 2A). However, only 29% of those genes are regulated in the WT hGRβ cell line, and the extent of gene regulation is blunted (visualized by the intensity of color). Moreover, many Diseases and Biological Functions were predicted to have opposing activation scores (Supplementary Figure 2B). The differences in predicted functions and signaling pathways were further evident when performing a comparison analysis of the top 15 canonical signaling pathways for each genotype by IPA (Figure 3b). The top eight canonical signaling pathways were similarly induced or repressed in the WT hGRβ- and G3134T hGRβ-expressing cells. However, the activation state of many of the other top canonical signaling pathways was predicted to occur in opposite directions. Differences in regulated functions and pathways were further substantiated using an independent functional annotation clustering analysis (Supplementary Table 1). Gene Ontology (GO) analysis found no common biological processes in the significantly regulated genes of WT hGRβ and G3134T hGRβ cells.
There were 1319 genes common to both genotypes and 99.6% of these genes demonstrated the same directionality of regulation. Not surprisingly, a comparison analysis of the commonly regulated genes revealed a high level of agreement for the top enriched functions and canonical pathways (Figure 3c). This suggested that the differences in enriched signaling pathways originated from the uniquely regulated genes in each genotype. A comparison analysis of the uniquely regulated genes revealed appreciable differences in the top Diseases and Biological Functions (Figure 3d). This was validated through GO analysis, where the only shared biological function between the two genotypes was the g-protein coupled receptor protein signaling pathway (Supplementary Table 2). Interestingly, the comparison analysis of the top canonical pathways in the unique gene sets indicated a similar direction of activation for the top five pathways, although there were differences in magnitude (Figure 3d). These data indicate similar pathways could be targeted through unique gene regulation. To independently confirm the expression changes in unique and commonly regulated genes, quantitative RT-PCR was performed on four additional mRNA samples from Uoff, WT hGRβ, and G3134T hGRβ cells (Figure 3e). Known GR target gene, Interleukin 13 receptor, alpha 2 (IL13RA2) was validated as uniquely regulated by WT hGRβ.19 Cerebellin 2 precursor (CBLN2) and TNF receptor-associated factor 1 (TRAF1) were validated as genes that demonstrate anticorrelated regulation (induced vs repressed) in WT hGRβ and G3134T hGRβ cells. Endothelin 3 (EDN3) validated as commonly repressed in both genotypes. Thus, the in vitro studies indicate that the G3134T polymorphism is capable of inducing gene expression changes that significantly alter signaling pathways that may underlie clinical phenotypes.
Glucocorticoid Responsiveness Differs in Participants Harboring the hGRβ G3134T Polymorphism
To determine whether the presence of the G3134T hGRβ polymorphism altered genome-wide gene expression in a physiological system, microarray analysis was performed on participants’ isolated monocytes differentiated to macrophages ex vivo, which express both hGRα and WT or G3134T hGRβ (Figure 4). Macrophages were treated for 6 h with vehicle (PBS) or 100 nM dexamethasone to assess the impact of the G3134T hGRβ polymorphism on glucocorticoid responsiveness. Gene lists were created in Partek Genomics Suite to identify significant differentially expressed genes. Analysis identified 900 significantly regulated genes in the vehicle-treated G3134T hGRβ macrophages compared to vehicle-treated WT hGRβ macrophages, indicating that the presence of the G3134T hGRβ polymorphism alters basal gene expression in primary human cells. Differences in the overall directionality of gene regulation were visualized by generating a heat map of significantly regulated genes and graphing induced vs repressed genes (Figure 4a). Compared to macrophages with WT hGRβ, macrophages homozygous for the G3134T hGRβ polymorphism preferentially induced 451 genes and repressed 449 genes. IPA predicted and graphed the relationship of the top canonical pathways related to genes with significantly different basal gene expression (Figure 4b). Several of the top canonical pathways shared similar biological functions. The top canonical pathways with overlapping functions were visualized with lines between each connected pathways (Figure 4b).
Glucocorticoid-regulated gene expression is altered by the presence of the G3134T hGRβ polymorphism in human macrophages. (a) mRNA isolated from seven WT hGRβ participants and three homozygous G3134T hGRβ participants were analyzed by the Agilent Whole Human Genome 4 × 44 multiplex format oligo array for gene expression. Heat map of significantly regulated gene probes by genotype was created with Partek Genomics Suite software and visually represent unique and commonly regulated targets. Red indicates induced genes and green indicates repressed genes. (b) The 25 top Canonical Signaling Pathways were visualized and shared functions identified by lines connecting nodes. The intensity of the red color is proportional to significance of P-value as determined by the Fisher’s Exact Test. (c) The number of probes statistically different following 6 h treatment with 100 nM dexamethasone compared to vehicle at P<0.05 with false discovery rate (step up) were sorted by Venn diagram. (d) All significant glucocorticoid-regulated genes were analyzed by IPA software. A comparison analysis was performed, and the top 15 Diseases and Biological Functions and Canonical Signaling Pathways are reported. The activation score is a prediction of the activation state of the molecules in a given function. Orange bars predict an overall increase in the activity of the pathway, while blue bars indicate a prediction of an overall decrease in activity. (e) Gene significantly regulated by glucocorticoids in both WT hGRβ participants and homozygous G3134T hGRβ participants were analyzed by IPA software. The top five Diseases and Biological Functions and Canonical Signaling Pathways are shown for each genotype. (f) A comparison analysis of the genes uniquely regulated by glucocorticoids in either WT hGRβ participants or participants genotyped as homozygous G3134T hGRβ was performed, and the top five Diseases and Biological Functions and Canonical Signaling Pathways are shown for each genotype.
In the presence of glucocorticoids, 705 genes were significantly regulated in macrophages from participants with WT hGRβ, and 936 genes were significantly regulated in macrophages from participants with the G3134T hGRβ polymorphism, with 302 genes common to both genotypes (Figure 4c). In addition to a greater total number of genes regulated by glucocorticoids in the participants with the G3134T hGRβ polymorphism, the ratio of induced to repressed genes shifted. Glucocorticoid treatment repressed 50.2% of regulated genes in participants with WT hGRβ and 71.7% of regulated genes in participants with the G3134T hGRβ polymorphism. The differences in ratio of repressed genes in response to glucocorticoid treatment resulted in opposing activation status for some of the top canonical signaling pathways as determined by IPA, although the activation status of the top 15 molecular and cellular functions was similar for both genotypes (Figure 4d). Gene ontology analysis identified several common biological functions, while four were unique to glucocorticoid regulation in participants with WT hGRβ (Organismal Physiological Process, Inflammatory Response, Response to Stimulus, and Negative Regulation of Cellular Physiological Process; Supplementary Table 3). There was high concordance for the direction of regulation for the genes common to both genotypes (301 of 302 genes regulated in the same direction in both genotypes), and the activation status of the predicted top functions and canonical signaling pathways was equivalent when analyzed by IPA (Figure 4e). Interestingly, the absence of the G3134T hGRβ polymorphism resulted in genes that were uniquely regulated by glucocorticoids only in macrophages from WT participants. Conversely, the presence of the G3134T hGRβ polymorphism resulted in the unique regulation of certain genes by glucocorticoids. A comparison analysis of the uniquely regulated genes predicted opposite activation status for all of the top five diseases and biological functions and canonical signaling pathways (Figure 4f). Thus, the ex vivo studies suggest that the presence of the G3134T hGRβ polymorphism alters both basal and glucocorticoid-responsive gene expression.
Discussion
The diversity in how an individual responds to both endogenous and exogenous glucocorticoids is determined in part by functional polymorphisms in hGR. However, the allele frequencies of known functional polymorphisms in the general population are relatively low.20, 21 In this study, we found that the G3134T hGRβ polymorphism is found at a much greater frequency than other well-characterized hGR polymorphisms. Moreover, we show that the G3134T hGRβ polymorphism is a new genetic factor influencing glucocorticoid sensitivity and signaling and clinical outcomes.
At the molecular level, glucocorticoid signaling is determined by ligand availability, GR expression levels, and GR sensitivity. Diminished sensitivity can lead to glucocorticoid resistance, which is characterized by hyperactivity of the hypothalamic–pituitary–adrenal axis and overproduction of adrenal hormones.21 The development of glucocorticoid resistance can lead to hypertension or chronic fatigue, and it limits the success of many glucocorticoid-based therapies.22 Polymorphisms located in the ligand-binding domain of GR are able to reduce ligand-binding affinity by 50%.23 hGR polymorphisms have also been described to reduce the transactivating capacity of GR, slow nuclear translocation, alter DNA binding, and disrupt coactivator interactions (reviewed in Quax et al24). For some polymorphisms, reduced transactivation is related to increased expression of the hGRβ variant.25 Our results show a significant increase in hGRβ levels in vitro, trends toward higher hGRβ levels in peripheral blood, and a corresponding increase in serum cortisol levels in participants carrying the G3134T hGRβ polymorphism. In response to the DST, participants carrying the G3134T hGRβ polymorphism demonstrated altered glucocorticoid sensitivity, and were unable to equivalently suppress serum cortisol compared to participants with WT hGRβ genotype. These data suggest that this polymorphism could contribute to changes in glucocorticoid sensitivity and consequently glucocorticoid resistance, and that within a population, the G3134T hGRβ polymorphism may contribute to diversity in glucocorticoid responsiveness between individuals and among races.
The presence of this polymorphism also results in changes to the inherent transcriptional activity of hGRβ. Whole-genome microarray analysis in U-2 OS cells, which lack hGRα but are stably transfected with WT or G3134T hGRβ, identified aberrant gene expression profiles when the G3134T variant was present. Independent validation of uniquely regulated genes suggests that the G3134T hGRβ polymorphism has the ability to selectively regulate genes that may be related to clinical pathology. The biological activities of IL13, a cytokine crucial to the development of allergic responses, are mediated through selective, high-affinity binding to IL13RA2, which induces rapid internalization.26 IL13RA2 has a protective role in the pathogenesis of allergic inflammation, and polymorphisms in the IL13 signaling pathway are associated with chronic respiratory diseases.27, 28 This gene was induced in U-2 OS cells in the presence of WT hGRβ but not G3134T hGRβ. TRAF1 also has an important role in the pathogenesis of allergic lung inflammation.29 Mice deficient in TRAF1 fail to elicit immune cell recruitment, proinflammatory cytokine expression, or airway hyper-responsiveness in an induced model of asthma. Interestingly, TRAF1 expression was repressed by WT hGRβ but induced when the G3134T polymorphism was present. Whether the IL13RA2 or TRAF1 genes are direct or indirect targets of hGRβ regulation is currently unknown. Both genes are directly regulated by tumor necrosis factor (TNF)-α, which also preferentially induces the expression of hGRβ over hGRα.30, 31, 32 Glucocorticoids are able to regulate TNF-α expression by antagonizing transactivation and destabilizing latent TNF-α.33, 34 Disruption of the feedback mechanisms necessary to maintain homeostasis by altered levels of hGRβ may lead to excessive or persistent inflammatory signaling.
In order to evaluate the impact of the G3134T hGRβ polymorphism on gene regulation in cells containing endogenous receptors, whole-genome microarray analysis was employed for peripheral blood macrophages from participants genotyped as WT or homozygous for the G3134T hGRβ polymorphism treated with and without dexamethasone. Similar to findings in stably transfected U-2 OS cells, the presence of the polymorphism uniquely altered basal gene expression. Compared to WT, macrophages from participants homozygous for the G3134T hGRβ polymorphism differentially expressed 900 genes. Gene ontology analysis indicated that these significantly regulated genes represented canonical pathways with overlapping functions, which primarily clustered into three nodes. The largest of these nodes comprised pathways related to immune cell activation and signaling. The microarray analysis also determined that primary cells from participants with the G3134T hGRβ polymorphism were differentially responsive to glucocorticoids. Although certain genes were commonly regulated between the two genotypes, macrophages from participants with the G3134T hGRβ polymorphism uniquely regulated 634 genes and lost the capacity to regulate 403 genes in response to glucocorticoid. These uniquely regulated genes resulted in the opposing predicted activation of signaling pathways related to the immune system, including PKCθ Signaling in T Lymphocytes, the Role of NFAT in Regulation of the Immune System, Tec Kinase Signaling, and the Production of Nitric Oxide and Reactive Oxygen Species. These data suggest that the G3134T hGRβ polymorphism may alter immune cell functions through dysregulation of both basal and glucocorticoid-responsive gene expression.
In the present study, we found an association between the G3134T hGRβ polymorphism and self-reported allergies, significantly different serum cortisol, and altered gene expression. While these data support an association, causation is not likely attributed to a single polymorphism. Strong linkage disequilibrium has been reported for several hGR polymorphisms, including G3134T hGRβ, indicating that hGR variants occur in individuals as haplotypes.8, 35 The presence of the Bcl1 polymorphism, which has been associated with elevated cortisol levels and shows linkage to G3134T hGRβ, may partly account for elevated cortisol levels in carriers and participants homozygous for G3134T.8, 36 Although in vitro data suggest that the G3134T hGRβ polymorphism is able to increase stability, it is not located in one of the 'AUUUA' motifs in the 3′ UTR known to confer mRNA stability. The A3669G hGRβ polymorphism does reside in this motif and demonstrates linkage disequilibrium with G3134T. This coupling could result in an additive effect on stability in patients with the G3134T hGRβ polymorphism. The absence of a reported metabolic phenotype in participants carrying the G3134T hGRβ polymorphism, which has been associated with the N363S polymorphism, may suggest an alternative hypothesis where certain haplotypes afford a benefit.18, 37 For some GR variants, a gene–environment interaction occurs.38 Stressful life events may predispose an individual to disease susceptibility when combined with genetic variants. The presence of the ER22/23EK or A3669G hGRβ polymorphism resulted in an increased risk of clinically relevant depressive symptoms in participants who had experienced childhood adversity.38 Lower birth weights have been predicted when maternal stress is coupled with the minor allele of the Tth111I hGR polymorphism.39 Moreover, the frequency of polymorphisms across genes varies greatly among ethnic populations, suggesting that the clinical response to any allele variant is a combination of gene, environment, and race.
Limitations of this study include the lack of comprehensive medical records and the inability to validate self-reported outcomes, which are inherently biased.40 Furthermore, the single clinical measurements limit our understanding of the exact effects of the G3134T hGRβ polymorphism on hormone kinetics or responsiveness. A parallel study utilizing the NIEHS EPR database and the self-reported survey found a 94% agreement between self-reported survey data and on-site medical history, which increases confidence in the results of the present study (personal correspondence). This study describes for the first time the prevalence of the G3134T hGRβ polymorphism in a large, genetically diverse population. Our findings indicate that this polymorphism regulates hGRβ signaling in immortalized cells and primary cells ex vivo, although the impact of the G3134T hGRβ polymorphism on glucocorticoid signaling in vivo is still unclear. However, these selective changes in gene expression identified in culture could be associated with the reported clinical outcomes. The identification of a polymorphism associated with a subset of inflammation-related transcripts suggests a regulatory mechanism that may be important to glucocorticoid therapy and the pathogenesis of glucocorticoid resistance.
References
Busillo JM, Cidlowski JA . The five Rs of glucocorticoid action during inflammation: ready, reinforce, repress, resolve, and restore. Trends Endocrinol Metab 2013;24:109–119.
Chrousos GP, Kino T . Glucocorticoid signaling in the cell. Expanding clinical implications to complex human behavioral and somatic disorders. Ann N Y Acad Sci 2009;1179:153–166.
Oakley RH, Sar M, Cidlowski JA . The human glucocorticoid receptor beta isoform. Expression, biochemical properties, and putative function. J Biol Chem 1996;271:9550–9559.
Oakley RH, Jewell CM, Yudt MR et al. The dominant negative activity of the human glucocorticoid receptor beta isoform. Specificity and mechanisms of action. J Biol Chem 1999;274:27857–27866.
Yudt MR, Jewell CM, Bienstock RJ et al. Molecular origins for the dominant negative function of human glucocorticoid receptor beta. Mol Cell Biol 2003;23:4319–4330.
He B, Cruz-Topete D, Oakley RH et al. Human glucocorticoid receptor beta regulates gluconeogenesis and inflammation in mouse liver. Mol Cell Biol 2015;36:714–730.
Charmandari E, Kino T, Souvatzoglou E et al. Natural glucocorticoid receptor mutants causing generalized glucocorticoid resistance: molecular genotype, genetic transmission, and clinical phenotype. J Clin Endocrinol Metab 2004;89:1939–1949.
Szczepankiewicz A, Leszczynska-Rodziewicz A, Pawlak J et al. Glucocorticoid receptor polymorphism is associated with major depression and predominance of depression in the course of bipolar disorder. J Affect Disord 2011;134:138–144.
Otte C, Wust S, Zhao S et al. Glucocorticoid receptor gene, low-grade inflammation, and heart failure: the Heart and Soul study. J Clin Endocrinol Metab 2010;95:2885–2891.
van Oosten MJ, Dolhain RJ, Koper JW et al. Polymorphisms in the glucocorticoid receptor gene that modulate glucocorticoid sensitivity are associated with rheumatoid arthritis. Arthritis Res Ther 2010;12:R159.
Koper JW, van Rossum EF, van den Akker EL . Glucocorticoid receptor polymorphisms and haplotypes and their expression in health and disease. Steroids 2014;92:62–73.
Derijk RH, Schaaf MJ, Turner G et al. A human glucocorticoid receptor gene variant that increases the stability of the glucocorticoid receptor beta-isoform mRNA is associated with rheumatoid arthritis. J Rheumatol 2001;28:2383–2388.
Varricchio L, Masselli E, Alfani E et al. The dominant negative beta isoform of the glucocorticoid receptor is uniquely expressed in erythroid cells expanded from polycythemia vera patients. Blood 2011;118:425–436.
Szczepankiewicz A, Rybakowski JK, Suwalska A et al. Glucocorticoid receptor polymorphism is associated with lithium response in bipolar patients. Neuro Endocrinol Lett 2011;32:545–551.
Spivey A . Environmental polymorphism registry. Environ Health Perspect 2006;114:A408–A409.
Lu NZ, Cidlowski JA . Translational regulatory mechanisms generate N-terminal glucocorticoid receptor isoforms with unique transcriptional target genes. Mol Cell 2005;18:331–342.
Jewell CM, Cidlowski JA . Molecular evidence for a link between the N363S glucocorticoid receptor polymorphism and altered gene expression. J Clin Endocrinol Metab 2007;92:3268–3277.
Jewell CM, Katen KS, Barber LM et al. Healthy glucocorticoid receptor N363S carriers dysregulate gene expression associated with metabolic syndrome. Am J Physiol Endocrinol Metab 2016;311:E741–E748.
Whirledge SD, Oakley RH, Myers PH et al. Uterine glucocorticoid receptors are critical for fertility in mice through control of embryo implantation and decidualization. Proc Natl Acad Sci USA 2015;112:15166–15171.
van Rossum EF, Koper JW, Huizenga NA et al. A polymorphism in the glucocorticoid receptor gene, which decreases sensitivity to glucocorticoids in vivo, is associated with low insulin and cholesterol levels. Diabetes 2002;51:3128–3134.
van den Akker EL, Russcher H, van Rossum EF et al. Glucocorticoid receptor polymorphism affects transrepression but not transactivation. J Clin Endocrinol Metab 2006;91:2800–2803.
Vingerhoeds AC, Thijssen JH, Schwarz F . Spontaneous hypercortisolism without Cushing's syndrome. J Clin Endocrinol Metab 1976;43:1128–1133.
Ruiz M, Lind U, Gafvels M et al. Characterization of two novel mutations in the glucocorticoid receptor gene in patients with primary cortisol resistance. Clin Endocrinol 2001;55:363–371.
Quax RA, Manenschijn L, Koper JW et al. Glucocorticoid sensitivity in health and disease. Nat Rev Endocrinol 2013;9:670–686.
Russcher H, Smit P, van Rossum EF et al. Strategies for the characterization of disorders in cortisol sensitivity. J Clin Endocrinol Metab 2006;91:694–701.
Kawakami K, Taguchi J, Murata T et al. The interleukin-13 receptor alpha2 chain: an essential component for binding and internalization but not for interleukin-13-induced signal transduction through the STAT6 pathway. Blood 2001;97:2673–2679.
Sivaprasad U, Warrier MR, Gibson AM et al. IL-13Ralpha2 has a protective role in a mouse model of cutaneous inflammation. J Immunol 2010;185:6802–6808.
Beghe B, Hall IP, Parker SG et al. Polymorphisms in IL13 pathway genes in asthma and chronic obstructive pulmonary disease. Allergy 2010;65:474–481.
Oyoshi MK, Bryce P, Goya S et al. TNF receptor-associated factor 1 expressed in resident lung cells is required for the development of allergic lung inflammation. J Immunol 2008;180:1878–1885.
David MD, Bertoglio J, Pierre J . Functional characterization of IL-13 receptor alpha2 gene promoter: a critical role of the transcription factor STAT6 for regulated expression. Oncogene 2003;22:3386–3394.
Pryhuber GS, Huyck HL, Staversky RJ et al. Tumor necrosis factor-alpha-induced lung cell expression of antiapoptotic genes TRAF1 and cIAP2. Am J Respir Cell Mol Biol 2000;22:150–156.
Webster JC, Oakley RH, Jewell CM et al. Proinflammatory cytokines regulate human glucocorticoid receptor gene expression and lead to the accumulation of the dominant negative beta isoform: a mechanism for the generation of glucocorticoid resistance. Proc Natl Acad Sci USA 2001;98:6865–6870.
Steer JH, Kroeger KM, Abraham LJ et al. Glucocorticoids suppress tumor necrosis factor-alpha expression by human monocytic THP-1 cells by suppressing transactivation through adjacent NF-kappa B and c-Jun-activating transcription factor-2 binding sites in the promoter. J Biol Chem 2000;275:18432–18440.
Smoak K, Cidlowski JA . Glucocorticoids regulate tristetraprolin synthesis and posttranscriptionally regulate tumor necrosis factor alpha inflammatory signaling. Mol Cell Biol 2006;26:9126–9135.
Mwinyi J, Wenger C, Eloranta JJ et al. Glucocorticoid receptor gene haplotype structure and steroid therapy outcome in IBD patients. World J Gastroenterol 2010;16:3888–3896.
Rosmond R, Chagnon YC, Holm G et al. A glucocorticoid receptor gene marker is associated with abdominal obesity, leptin, and dysregulation of the hypothalamic-pituitary-adrenal axis. Obes Res 2000;8:211–218.
Majer-Lobodzinska A, Adamiec-Mroczek J . Glucocorticoid receptor polymorphism in obesity and glucose homeostasis. Adv Clin Exp Med 2017;26:143–148.
Bet PM, Penninx BW, Bochdanovits Z et al. Glucocorticoid receptor gene polymorphisms and childhood adversity are associated with depression: New evidence for a gene-environment interaction. Am J Med Genet B Neuropsychiatr Genet 2009;150B:660–669.
Mparmpakas D, Zachariades E, Sotiriadis G et al. Differential expression of placental glucocorticoid receptors and growth arrest-specific transcript 5 in term and preterm pregnancies: evidence for involvement of maternal stress. Obstet Gynecol Int 2014;2014:239278.
Ritter PL, Stewart AL, Kaymaz H et al. Self-reports of health care utilization compared to provider records. J Clin Epidemiol 2001;54:136–141.
Acknowledgements
This work was supported by the Intramural Research Program of the National Institutes of Health, National Institute of Environmental Health Sciences.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Laboratory Investigation website .
Polymorphisms in the glucocorticoid receptor (GR) have been associated with altered glucocorticoid sensitivity and disease. In this study, the authors demonstrate that the G3134T hGRβ polymorphism is associated with race and shows linkage disequilibrium with other GR polymorphisms. The G3134T hGRβ polymorphism is also associated with self-reported allergies, significant differences in serum cortisol levels, and altered gene expression.
Rights and permissions
About this article
Cite this article
Whirledge, S., Jewell, C., Barber, L. et al. Generating diversity in human glucocorticoid signaling through a racially diverse polymorphism in the beta isoform of the glucocorticoid receptor. Lab Invest 97, 1282–1295 (2017). https://doi.org/10.1038/labinvest.2017.76
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/labinvest.2017.76