Abstract
Part of developing therapeutics is the need to identify patients who will respond to treatment. For HER2-targeted therapies, such as trastuzumab, the expression level of HER2 is used to identify patients likely to receive benefit from therapy. Currently, chromogenic immunohistochemistry on patient tumor tissue is one of the methodologies used to assess the expression level of HER2 to determine eligibility for trastuzumab. However, chromogenic staining is fraught with serious drawbacks that influence scoring, which is additionally flawed due to the subjective nature of human/pathologist bias. Thus, to advance drug development and precision medicine, there is a need to develop technologies that are more objective and quantitative through the collection and integration of larger data sets. In proof of concept experiments, we show multiplexed ion beam imaging (MIBI), a novel imaging technology, can quantitate HER2 expression on breast carcinoma tissue with known HER2 status and those values correlate with pathologist-determined IHC scores. The same type of quantitative analysis using the mean pixel value of five individual cells and total pixel count of the entire image was extended to a blinded study of breast carcinoma samples of unknown HER2 scores. Here, a strong correlation between quantitation of HER2 by MIBI analysis and pathologist-derived HER2 IHC score was identified. In addition, a comparison between MIBI analysis and immunofluorescence-based automated quantitative analysis (AQUA) technology, an industry-accepted quantitation system, showed strong correlation in the same blind study. Further comparison of the two systems determined MIBI was comparable to AQUA analysis when evaluated against pathologist-determined scores. Using HER2 as a model, these data show MIBI analysis can quantitate protein expression with greater sensitivity and objectivity compared to standard pathologist interpretation, demonstrating its potential as a technology capable of advancing cancer and patient diagnostics.
Similar content being viewed by others
Main
Methods to accurately characterize the prevalence, location, and expression level of relevant biomarkers are critical for the success of targeted therapies. In the example of HER2-targeted therapies, such as trastuzumab (Herceptin), HER2 expression is predictive for efficacy through semi-quantitative assessment of patient tumor samples by chromogenic immunohistochemistry (IHC) or fluorescent in situ hybridization.1 However, only 20–30% of HER2-positive patients will respond to treatment.2, 3 While this has made a tremendous impact on HER2-positive cancer diagnosis, more sensitive and objective diagnostic methods can potentially better identify patients who will benefit from targeted therapies.
An integral part of developing future diagnostics is the capability to fully quantitate and visualize multiple markers simultaneously on tissue sections. Building upon semi-quantitative single marker assessment, like HER2, will be full quantitation of the expression level of interrogated proteins in the context of other markers through multiplexing. This will provide an in-depth analysis of attributes to better recognize ideal patients for developing drugs and evaluation. As mentioned above, only 20–30% of HER2-positive patients will respond to trastuzumab, suggesting a singleplex is not sufficient to identify the most ideal patients.2, 3 There is also anecdotal evidence for all anti-PD-1 or anti-PD-L1 therapeutics that patients negative for these markers by IHC respond to treatment, indicating again that multiple markers and quantitation will be needed to capture ideal responders. Similarly, data from quantitation of multiple markers will afford a better evaluation of the patient to identify the best course of treatment. Therefore, new technologies should permit: (1) quantitation of signal and (2) multiplexing to generate large, in-depth data sets.
To this end, multiplexed ion beam imaging (MIBI) of tissue sections stained with metal-labeled antibodies represents a new technology that can potentially satisfy the quantitation and multiplexing requirements described above.4, 5 Here, stained tissue sections on conductive surfaces are scanned with a beam of energy that ionizes the metals on the antibody to generate secondary ions that can be detected by a imaging mass spectrometer.4, 5 This type of analysis is capable of expanded multiplexing since fluorescent spectral overlap no longer constrains the number of markers available as distinct non-overlapping secondary ions signatures from the metals are detected.5 Similarly, the staining process for multiplexing is more efficient because the primary antibody is directly conjugated to the metal and does not require the amplification steps found in other multiplexing systems.4 MIBI analysis also records the magnitude of the secondary ions that are emitted, which is a direct correlation with target expression level.4 The quantitative values, such as mean pixel value and total pixel count, for expression level of protein markers can be collected from the rasters generated from this analysis with the aid of imaging software.
In the HER2-targeted therapy example, chromogenic IHC with diaminobenzidine (DAB) is first performed using FDA-approved companion diagnostics, followed by pathologist evaluation for HER2 staining intensity and percent of HER2-positive cells to determine eligibility for treatment.6 Trained pathologists then use a semi-quantitative four-tier scoring system from 0 to 3+ to assess samples, in which only 3+ cases qualify for the medicines.6
However, this process and methodology has many flaws that affect scoring and possibly the decision to provide treatment. First, manual scoring by a pathologist is subjective in nature and semi-quantitative, which has led to inter-observer discordance in multiple cancer types, including gastroesophageal adenocarcinoma and gastric cancer.7, 8, 9, 10, 11 Second, the DAB detection technology is limiting as the dynamic range of DAB staining is severely restricted to one to two orders of magnitude due to the biophysical properties of DAB that block 99% of light on optimally stained samples.12 In addition, the unpredictable and variable nature of DAB detection technology has been associated with scoring discrepancies between facilities and institutions.13, 14, 15, 16 Together, this type of assessment illustrates a need to eliminate subjective and semi-quantitative scoring by pathologists, and to develop technologies that may be more objective and quantitative.
Given these subjective and technical issues associated with current diagnostics, MIBI analysis is uniquely poised to both resolve these issues and advance diagnostics through its technical capabilities to expand multiplexing and quantitate signals. Indeed, recent studies have demonstrated these capabilities by assessing at least seven markers in breast carcinoma tissue.4 However, an in-depth assessment of the quantitative capability of MIBI analysis was not performed. Therefore, our current studies represent proof of concept to validate MIBI analysis as a technology that can be used to quantitate protein expression level on tissue sections without bias. Using the clinically relevant HER2 scoring system in breast carcinoma as a model, we demonstrate MIBI analysis can quantitate HER2 protein expression level on breast carcinoma tissue. Here, we show the metal-labeled HER2 antibody discriminates between different expression levels of HER2 by MIBI analysis, and quantitation of MIBI-analyzed rasters strongly correlate with the known HER2 IHC status of cell line pellets and breast carcinoma samples. To extend these findings, we performed a blinded analysis of breast carcinoma samples and observed that quantitation by MIBI analysis strongly correlates with quantitation derived from the fluorescence-based automated quantitative analysis (AQUA) system and pathologist-determined IHC score. In summary, MIBI analysis represents a promising new technology that can objectively score protein expression level that will aid in the advancement of diagnostics and drug development.
Materials and methods
Conjugation of Metal-Chelating Polymer to Antibodies
HER2 (clone D8F12, Cell Signaling Technologies) and dsDNA (clone HYB 331-01, Abcam) antibody were labeled with Maxpar X8 Reagents (Fluidigm) according to manufacturer’s protocol. Briefly, Holmium 165 (for HER2) or Samarium 154 (for dsDNA) was chelated to polymer, while antibodies were partially reduced with TCEP.17, 18 Polymers chelated with metal were then incubated with reduced antibodies for conjugation and subsequently washed.17, 18 Concentration of metal-labeled antibodies was determined by A280 absorbance and resuspended to 200 μg/ml with PBS antibody stabilizer (Candor).
Immunohistochemistry
Freshly cut 4 μm sections of formalin fixed paraffin embedded (FFPE) cell line pellets and breast carcinoma tissue with known HER2 score were stained for HER2 antibody (Cell Signaling Technology; clone D8F12) at 0.3 μg/ml on a Ventana Benchmark XT platform using CC1 mild antigen retrieval and UltraView Universal DAB detection (Ventana). Stained slides were scanned and images were taken using gslideViewer software (Genentech).
IHC with Polymer-Conjugated Antibodies
Freshly cut 4 μm sections of a tissue microarray (TMA) consisting of FFPE cell line pellets and breast carcinoma tissue cores with known HER2 score were placed on a silicon wafer (Microsurfaces) treated with Vectabond (Vector Laboratories). TMAs were baked for 1 h at 65 °C, deparaffinized, and rehydrated. Target antigen retrieval (DAKO) was performed for 20 min at 99 °C and tissues were blocked in 10% donkey serum/TBST. HER2-165Ho and dsDNA-154Sm antibody were cocktailed at 10 μg/ml each in blocking buffer and incubated overnight at 4 °C in a humidity chamber. After 18 h, slides were post-fixed in 2% glutaraldehyde/PBS and counterstained with Mayer’s hematoxylin for 1 min. Stained sections were stored in a vacuum chamber until MIBI analysis.
MIBI Analysis
MIBI analysis was performed on the CAMECA NanoSIMS 50L at the Stanford Nano Shared Facilities. The O− primary ion beam was used to enhance the positive secondary ion yields for the species of interest. Control spots with HER2-165Ho and dsDNA-154Sm antibody were used to tune to the proper spectral peaks and optimize secondary ion detection. Optimization of secondary ion optics was performed for each sample. Beam spot size was ~0.5 μm at the D1_2 aperture setting used for all samples. Images of 256 × 256 pixels were acquired from a field of view of 100 × 100 μm. The ion beam dwell time was 1 ms per pixel and 25 repeat scans were taken for each analysis area. Each image took ~27 min to acquire using the NanoSIMS instrument. Images presented were pseudo-colored using ImageJ (NIH).
Quantitation of MIBI Analysis
Rasters from MIBI analysis were quantitated using mean pixel value of five cells or total pixel count of the entire image with the aid of ImageJ imaging software. For mean pixel value, the 25 repeated scans were summed and five regions of interest (cells) were chosen from that image that did not overlap with other cells and were entirely in the field of view based on dsDNA/hematoxylin-merged images. Traces were drawn around dsDNA/hematoxylin border. Mean pixel values were obtained for each cell using ImageJ. Each sample contains a metafile that contains the primary ion beam current measured after MIBI analysis, which is the amount of O− beam current to the sample during the analysis. Mean pixel value was normalized to primary beam current to permit comparison from one sample to another. ImageJ was also used to obtain the total pixel count for HER2 staining in the entire image, which was then normalized to primary beam current to permit comparison from one sample to another. Student’s t-test for statistical significance and Spearman correlation analysis for strength of correlation was performed using Graphpad Prism.
HER2 Blinded Study
To construct a TMA composed of invasive ductal breast adenocarcinoma and cell line pellets for the blind study, cases of breast carcinoma tissue samples with unknown HER2 status were first stained chromogenically for HER2 as described above. A trained pathologist scored all cases and tumor areas that represented 0–3+ HER2 scores were circled and cored with the tumor area in the center. For generating the cell line pellet cores, existing FFPE blocks of cell lines with known HER2 status were chosen and punched. Breast carcinoma tissue and cell line pellet cores were then randomly positioned to construct the TMA. HER2 IHC scores were confirmed on the TMA using chromogenic staining by the same pathologist who scored the original cases. The pathologist then generated a key for the location of each sample on the TMA. For AQUA quantitation, the randomized TMA was deparaffinized and rehydrated, Target antigen (DAKO) retrieved for 20 min at 99 °C, and transferred onto a DAKO autostainer platform. TMA was then treated with 3% H2O2, incubated with HER2 antibody (Cell Signaling Technology; clone D8F12) at 0.3 μg/ml for 60 min at RT, stained with Powervision anti-rabbit HRP detection (DAKO) and TSA Alexafluor 488 (Life technologies), counterstained with DAPI, and mounted in Prolong Gold mounting media. IF-stained TMA underwent AQUA software analysis at Genoptix. Image acquisition for AQUA analysis was ~1 min per core. For MIBI quantitation, the randomized TMA was stained with metal polymer-conjugated antibodies as described above. The center of the core underwent MIBI analysis, which took ~27 min per core, and quantitation of HER2 staining was performed as described above by an individual with no knowledge of core locations and their corresponding IHC and AQUA scores. After analysis, samples were unblinded, and MIBI quantitation, AQUA data, and pathologists-derived IHC score were compared. Student’s t-test for statistical significance and Spearman correlation analysis for strength of correlation was performed using Graphpad Prism.
Results
Metal-Chelating Polymer-Modified Antibodies Retain Ability to Discriminate HER2 Expression Levels
MIBI relies on the detection of secondary ions emitted after bombardment of elemental metal by an energy source.4, 5 Therefore, primary antibodies are modified with a polymer that chelates metal ions to be detected by the NanoSIMS mass-analyzing instrument. The antibodies used in our studies were modified using the Fluidigm Maxpar X8 conjugation kit in which the antibody was partially reduced to generate free thiols that react with the polymer to create a metal-chelating polymer-conjugated antibody.17, 18
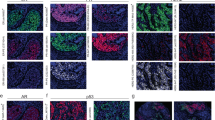
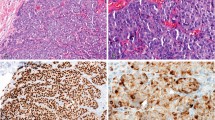
The HER2 antibody used in our studies was clone D8F12 from Cell Signaling Technologies, which is a rabbit monoclonal antibody that has the sensitivity to discriminate between various expression levels of HER2. In recent studies, the D8F12 antibody demonstrated similar sensitivity and specificity with other FDA-approved antibodies including A0485 (rabbit polyclonal, Dako) and CB11 (mouse monoclonal, Ventana).19 Because conjugating the polymer to antibodies involves the reduction of interchain disulfide bonds that may alter recognition of the antigen,17, 18 studies were undertaken to ensure that conjugation process did not alter D8F12’s ability to discriminate between HER2 expression levels. To this end, DAB staining with unmodified and modified D8F12 as the primary antibody was performed on cell lines and human invasive ductal breast carcinoma with different levels of HER2 expression. As shown in Figure 1, polymer-conjugated D8F12 discriminated HER2 expression levels similarly to unmodified D8F12 on cell line pellets (Figures 1a–d) and breast carcinoma tissue samples (Figures 1e–h). These data suggest that conjugating the metal-chelating polymer to D8F12 did not alter its ability to discriminate between different HER2 expression levels.
HER2 antibody clone D8F12 modified with metal-chelating polymers retains ability to discriminate HER2 expression levels on cell pellets and breast carcinoma tissue. Cell line pellets and breast carcinoma tissue with different HER2 expression levels were stained with unmodified or polymer-modified HER2 antibody clone D8F12, and visualized with DAB using the Ventana UltraView Universal DAB detection system. (a–d) Chromogenic staining of cell line pellets with known expression levels of HER2 with metal-chelating polymer labeled (top row) and unmodified (bottom row) D8F12. (e–h) Chromogenic staining of breast carcinoma tissue with known HER2 status using metal-chelating polymer labeled (top row) and unmodified (bottom row) D8F12.
In addition to the ability to discriminate between various HER2 expression levels, the HER2 expression pattern and staining intensity was similar between the unmodified and polymer-modified antibody. In control cell line pellets, the 0 score cell line exhibited little to no staining (Figure 1a), while the 1+ cell line showed comparable weak cytoplasmic HER2 staining (Figure 1b) with both antibodies at the same concentration. Similarly, the 2+ cell line showed modest membranous staining (Figure 1c), while the 3+ cell line exhibited darker membranous staining (Figure 1d) between the unmodified and polymer-modified antibody. In the breast carcinoma tissue, the 0 and 1+ cases showed no staining or weak cytoplasmic staining (Figures 1e and f), respectively. Finally, the 2+ and 3+ cases exhibited similar membranous staining patterns and intensities with both antibodies (Figures 1g and h). Together, these data demonstrate that conjugation of the polymer to D8F12 did not alter sensitivity and specificity of the antibody.
Quantitation of HER2 Expression by MIBI Analysis of Cell line Pellets and Breast Carcinoma Tissue Correlates with Pathologist-Determined HER2 IHC Score
Because the polymer-modified antibody still retained the ability to discriminate between HER2 expression levels by IHC, studies were performed to determine if quantitation of MIBI-analyzed samples correlated with pathologist-determined HER2 IHC score. To this end, a TMA that included cell line pellets and human invasive ductal breast carcinoma samples with known HER2 IHC scores was constructed and stained with metal-labeled antibodies against dsDNA and HER2, and counterstained with hematoxylin. As shown in Figure 2, differences in HER2 staining can be visually observed in different cell lines (Figure 2a) and breast carcinoma samples (Figure 2b), suggesting that the metal-labeled HER2 antibody and MIBI analysis can be qualitatively used to visually discriminate samples that express different levels of HER2. Similar to chromogenic staining, membranous staining is also observed in 3+ cell line pellets (Figure 2a, right) and breast carcinoma samples (Figure 2b, right, and Supplementary Figures S1A–D). There is some overlap of HER2 and dsDNA/hematoxylin staining in some cells with MIBI analysis; however, this observation is also seen with chromogenic staining (Figure 2 and Supplementary Figures S1A–E).
MIBI imaging analysis of metal-labeled HER2 antibody stained cell pellets and breast carcinoma tissue samples expressing various levels of HER2. Cell line pellets and breast carcinoma tissue with different HER2 expression levels were stained with HER2-165Ho antibody clone D8F12 (green), dsDNA-154Sm antibody (red), and hematoxylin (blue). The samples underwent MIBI analysis using the CAMECA NanoSIMS 50L and pseudocoloring with ImageJ. (a,b) HER2, dsDNA, and hematoxylin-merged rasters from MIBI analysis of cell line pellets (a) and breast carcinoma tissue (b) expressing different levels of HER2. MIBI rasters are 50 × 50 μm.
More importantly, the rasters from MIBI analysis can be used to obtain the mean pixel value of the marker of interest to objectively quantitate expression level. To determine if the mean pixel value of HER2 staining correlated with the pathologist-determined HER2 IHC score, five regions of interest (that is, cells) were identified and demarcated based on the merged dsDNA/hematoxylin images using ImageJ. The mean pixel value of HER2 staining within those cells was compiled and normalized to the amount of current that was passed through the sample during MIBI analysis. As shown in Figure 3a, a perfect Spearman correlation coefficient (rs=1) was observed between the average of the normalized HER2 mean pixel value of the five chosen cells and pathologist-determined score (Figure 3a).
Quantitation of HER2 expression by MIBI analysis of cell line pellets and breast carcinoma tissue correlates with pathologist-determined HER2 IHC score. (a, b) The mean pixel value of HER2 staining for five cells from each sample were obtained and normalized to the current passed through the sample. The normalized mean pixel value for each cell from cell line pellets (a) and the tumor region of breast carcinoma samples (b) was plotted against their known HER2 IHC score. Each symbol represents the normalized mean pixel value of one cell and the line represents the average of those five cells. Asterisks represent statistical significance based on two-tailed unpaired Student’s t-test between denoted samples (*P<0.05; **P<0.01; ***P<0.001). (c, d) The HER2 total pixel count for each sample was also obtained and normalized to the current passed through the sample. The normalized total pixel count for cell line pellets (c) and breast carcinoma samples (d) were plotted against their known HER2 IHC score.
A similar methodology was used to determine if there was a correlation between the mean pixel value of HER2 staining and pathologist-given IHC score on breast carcinoma samples. Like the data from the cell line pellets, there was a perfect Spearman correlation coefficient (rs=1) between the pathologist-determined score and normalized average mean pixel value of HER2 staining (Figure 3b) for five tumor cells.
However, using five arbitrarily chosen cells may not be representative of the entire image. Therefore, the total pixel count for the entire image of a sample was obtained using ImageJ and compared to the IHC score. Like mean pixel value of five individual cells, the normalized total pixel count increased with increasing IHC scoring tier in both cell line pellets (Figure 3c) and breast carcinoma samples (Figure 3d).
HER2 Quantitation from MIBI Analysis Strongly Correlates with Pathologist-Given Score and AQUA-Based Quantitation in Blind Study of Breast Carcinoma Samples
In the previous proof of concept experiment, the HER2 status of the cell line and breast carcinoma tissue sample was known before MIBI analysis. However, quantitation based on MIBI analysis must be able to confidently and reliably score markers withouta priori scoring knowledge if MIBI-based marker analysis is to be used in a diagnostic setting. Therefore, a blind study was undertaken to determine if quantitation by MIBI analysis correlated with pathologist-determined IHC score on human invasive ductal breast carcinoma samples of unknown HER2 status. Quantitative immunofluorescence by AQUA software, an industry-accepted application, was also included in the blind study to compare the performance of MIBI analysis against AQUA technology.20 To this end, MIBI quantitation, AQUA quantitation, and pathologist scoring were performed on breast carcinoma tissue cores of unknown HER2 status and cell line pellets that were randomly positioned on a TMA.
After staining with the appropriate antibodies and analysis by the different quantitation systems, MIBI-based quantitation of HER2 staining from five cells of the cell line pellets were averaged and compared to the pathologist-determined IHC score. As shown in Figure 4a, there was a very strong and significant correlation between normalized mean pixel value and IHC score in cell line pellets (Figure 4a; rs=0.8807, P<0.0001, 95% CI=0.7035–0.9548). Similarly, the normalized average mean pixel value of HER2 staining from five cells within the tumor area for each breast carcinoma tissue core was compared to the IHC score. Like the cell pellet data, there was a very strong and significant correlation between the normalized average mean pixel value from MIBI analysis and IHC score on breast carcinoma tissue samples (Figure 4b; rs=0.8310, P<0.0001, 95% CI=0.6830–0.9135). Further, the difference in the mean pixel value between adjacent HER2 scoring tiers was significantly different for the breast carcinoma samples (Figure 4b).
HER2 quantitation based on mean pixel value strongly correlates with AQUA quantitation and pathologist-given score in blind study of human invasive ductal breast carcinoma samples. The mean pixel value of HER2 staining was obtained for five cells from each sample, normalized to the final amount of current passed through the sample, and averaged to obtain average normalized mean pixel value. AQUA values and pathologist-given IHC score were unblinded after MIBI analysis of all samples. (a, b) The average normalized mean pixel value from each cell line pellet (a;rs=0.8807, P<0.0001, 95% CI=0.6487–0.9484) and breast carcinoma sample (b; rs=0.8310, P<given IHC score) was plotted against its corresponding pathologist-determined IHC score. (c, d) The AQUA value for each cell line pellet (c; rs=0.8701, P<0.0001, 95% CI=0.6709–0.9522) and breast carcinoma sample (d; rs=0.8313, P<0.0001, 95% CI=0.6835–0.9137) was plotted against its corresponding pathologist-determined IHC score. (e,f) The average normalized mean pixel value from each cell line pellet (e; rs=0.8452, P<0.0001, 95% CI=0.6156–0.9425) and breast carcinoma sample (f; rs=0.8437, P<0.0001, 95% CI=0.7049–0.9203) was plotted against its corresponding AQUA value. Each symbol represents the normalized average mean pixel value of five cells from the sample. Line represents the mean of all samples in each HER2 tier. rs represents the Spearman correlation coefficient. P represents statistical significance of Spearman correlation coefficient (****P<0.0001). Asterisks represent statistical significance based on two-tailed unpaired Student’s t-test between denoted samples (*P<0.05; **P<0.01). Note: there is an additional data point for the IHC score 0 in the MIBI vs IHC graph compared to the AQUA vs IHC graph as AQUA data were not obtained for that corresponding data point.
Fluorescence values from AQUA analysis were also compared against pathologist-determined IHC score to assess the strength of the correlation between the two systems. Similar to MIBI quantitation, there was a very strong and significant correlation between AQUA quantitation and IHC score in cell line pellets (Figure 4c; rs=0.8701, P<0.0001, 95% CI=0.6709–0.9522) and breast carcinoma tissue (Figure 4d; rs=0.8313, P<0.0001, 95% CI=0.6835–0.9137).
Analysis was also performed to determine the strength of correlation between normalized mean pixel value from MIBI analysis and quantitation from AQUA software analysis. As shown in Figures 4e and f, there was a very strong and significant correlation between the normalized mean pixel value from MIBI analysis and fluorescence units from AQUA analysis in cell line pellets (Figure 4e; rs=0.8452, P<0.0001, 95% CI=0.6156–0.9425) and breast carcinoma tissue samples (Figure 4f; rs=0.8437, P<0.0001, 95% CI=0.7049–0.9203), suggesting that there is strong agreement in the quantitative analysis of the two systems.
To again ensure that using the mean pixel value of five cells was representative of the sample, correlation analysis was also performed using total pixel count of the entire image. Like the mean pixel value analysis, normalized total pixel count strongly correlated with IHC score in cell line pellets (Figure 5a; rs=0.8770, P<0.0001, 95% CI=0.6952–0.9533) and breast carcinoma samples (Figure 5b; rs=0.8139, P<0.0001, 95% CI=0.6537–0.9043). Further analysis between normalized pixel count and AQUA score also showed strong correlation in cell lines (Figure 5c; rs=0.8906, P<0.0001, 95% CI=0.7181–0.9600) and breast carcinoma samples (Figure 5d; rs=0.8339, P<0.0001, 95% CI=0.6879–0.9150).
HER2 quantitation based on total pixel count strongly correlates with AQUA quantitation and pathologist-given score in the blind study of human invasive ductal breast carcinoma samples. The HER2 total pixel count was obtained from each sample and normalized to the final amount of current passed through the sample. (a, b) The total pixel count from each cell line pellet (a; rs=0.8602, P<0.0001, 95% CI=0.6487–0.9484) and breast carcinoma sample (b; rs=0.8310, P<given IHC score) was plotted against its corresponding pathologist-determined IHC score. (c,d) The average normalized total pixel count from each cell line pellet (c; rs=0.8452, P<0.0001, 95% CI=0.6156–0.9425) and breast carcinoma sample (d; rs=0.8437, P<0.0001, 95% CI=0.7049–0.9203) was plotted against its corresponding AQUA value. Each symbol represents one sample. Line represents the mean of all samples in each HER2 tier. rs represents the Spearman correlation coefficient. P represents statistical significance of Spearman correlation coefficient (****P<0.0001).
Comparison of correlation coefficients between normalized total pixel count and mean pixel value was also performed to assess if the two quantitation parameters gave similar results. Indeed, the correlation coefficients were comparable between the two parameters in the MIBI vs IHC (Table 1) and MIBI vs AQUA analysis (Table 1) for the cell line pellets. In breast carcinoma samples, the correlation values from mean pixel value and total sum pixels analysis were also similar in the two comparisons (Table 1), suggesting quantitative analysis using the two parameters may be used interchangeably.
To evaluate the performance of MIBI versus AQUA, the correlation coefficient from the two systems and their corresponding IHC score was compared. When comparing using the mean pixel value parameter, the correlation coefficients were quite similar with cell line pellets (Table 2) and breast carcinoma samples (Table 2). Comparison of the MIBI vs IHC and AQUA vs IHC correlation values using the total pixel count parameter also gave similar results in cell line pellets (Table 3) and breast carcinoma (Table 3). Together, these data suggest MIBI quantitation is on par with AQUA quantitation.
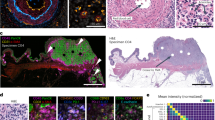
Finally, to assess if the MIBI-analyzed rasters visually corresponded with each IHC scoring tier and AQUA analysis, samples with a normalized mean pixel value similar to the average within each IHC tier was identified. As shown in Figure 6, differences in HER2 expression level could be visually discriminated from the rasters of those identified samples as well as their corresponding chromogen (Figure 6b) and immunofluorescent (Figure 6c) images, demonstrating visual correlation and confirmation of HER2 stratification.
MIBI-analyzed rasters visually correlate with IHC scores and immunofluorescent (IF) images of breast carcinoma samples from blind study. Briefly, breast carcinoma samples from blind study were stained with HER2 (gray) and DAPI (not shown) for AQUA analysis. Serial section of TMA was stained with HER2-165Ho antibody clone D8F12 (green), dsDNA-154Sm antibody (red), and hematoxylin (blue) for MIBI analysis. The average normalized mean pixel value was calculated for each IHC scoring tier, and a sample with a similar value was identified. (a–c) The MIBI-analyzed raster (a), chromogen image (b), and IF image (c) for identified samples are shown. The average normalized mean pixel value for each tier is listed (a), as are the normalized mean pixel value (a), IHC score (b), and AQUA values (c) for identified samples. MIBI rasters are 100 × 100 μm. Scale bar for chromogen images are 100 μm as indicated in image. Scale bar for IF images are 100 μm as indicated in image.
Discussion
For HER2-targeted therapies, such as Herceptin, the expression level and location of HER2 is predictive of response.1 As such, a scoring system based on HER2 IHC staining is used to determine which patients should receive therapy.6 While undeniably influential for the treatment of HER2-positive cancers, scoring discordance between different pathologists and institutions have been reported in multiple cancer types; thus highlighting the technical and subjectivity issues that are inherent in IHC analysis.7, 8, 9, 10, 11, 13, 14, 15, 16 Furthermore, there is a need for technologies that can quantitate markers, and multiplex simultaneously to help identify patient populations that will benefit from therapeutics and make more informed choices in the treatment of patients.21
One such technology is MIBI in which target detection reagents, such as antibodies, are conjugated to metal-chelating polymers and analyzed with an imaging mass spectrometer.4, 5 Recent studies have demonstrated the expanded multiplexing capabilities of mass spectrometry-based imaging to at least 32 markers.4, 22 However, a comprehensive assessment of its quantitative ability for expression level has not been performed. Therefore, these studies represent proof of concept for those capabilities using the clinically relevant HER2 scoring system. We first showed that conjugation of HER2 antibody clone to the metal-chelating polymer had no effect on target antigen recognition (Figures 1 and 2) because the process includes partial reduction of interchain disulfide bonds that could negatively impact antigen binding.17, 18 We next determined that MIBI-analyzed samples with known HER2 status could be used to quantitate HER2 with ImageJ in which the normalized mean pixel value of five cells or total pixel count correlated with pathologist-given IHC scores (Figure 3). To extend those findings, we performed a blind study to compare MIBI versus AQUA quantitation, an industry-accepted quantitation technology, and pathologist-given scores on human invasive ductal breast carcinoma samples with unknown HER2 status. Similar to the results with known IHC score samples, MIBI quantitation, based on mean pixel value and total pixel count, strongly correlated with pathologist-given IHC score and AQUA quantitation (Figures 4 and 5) with samples of previously unknown HER2 IHC score. In addition, the correlation coefficients between mean pixel values and total pixel count were similar (Table 1), suggesting the parameters may be used interchangeably. Finally, the correlation coefficient between MIBI-based and AQUA quantitation versus pathologist score was very similar (Tables 2 and 3), suggesting that MIBI-based quantitation was comparable to AQUA quantitation. Therefore, using HER2 in breast carcinoma as a model, these data indicate that MIBI-based analysis can be used to measure protein expression level in tissue sections.
Interestingly, the correlation of MIBI vs AQUA quantitation in the blind study appears to have a non-linear relationship (Figures 4 and 5) due to the plateauing MIBI signal of few samples observed at the higher expression range. This may be attributed to the inclusion of normal cells in the analysis since the multicell type breast tissue analysis includes them, while this is irrelevant for the homogeneous cell line pellets, whose relationship is linear with AQUA quantitation. Indeed, some breast sample images contain normal cells (Figure 2b, far right; Figure 6a, far right; Supplementary Figures S1A, C and D), which influences the analysis by reducing the MIBI quantitation values. Although all breast tissue analysis are impacted by the inclusion of normal cells, analysis of 3+ HER2 samples are influenced greater because the difference in HER2 signal between tumor cells and non-tumor cells is larger than the difference between HER2 signal for tumor cells and non-tumor cells in 0, 1+, or 2+ tissue. Exclusion of non-tumor cells from the MIBI analysis area would be ideal, but technically difficult and challenging; therefore, careful attention was paid to coring and imaging tumor areas.
MIBI analysis offers a few quantitative advances over standard chromogenic IHC. First, MIBI analysis removes issues that limit quantitation associated with DAB staining.12 In contrast to DAB staining, which has one to two orders of magnitude,12 MIBI has a quantitation range of five orders of magnitude for higher sensitivity and larger range of detection.4, 5 Second, MIBI removes the subjectivity of pathologist-based semi-quantitative assessment. With MIBI analysis, a digital image is generated in which quantitative values can be extracted with imaging software to obtain a continuous scale of values associated with expression level. This prevents the subjective situation in which pathologists are asked to score samples that may sit at the border of the scoring tiers. Similarly, an objective scoring system would also reduce discordance between institutions and facilities. Together with our validation studies, MIBI analysis removes issues that limit quantitation, semi-quantitative assessment, and pathologist subjectivity to score samples more quantitatively and objectively.
To better diagnose individuals and capture patients that will respond to treatment, there is a need for an in situ protein analytic method that can provide multiple data points. MIBI analysis is well positioned to afford that information since it can provide protein expression data for multiple markers and spatial information to interrogate where proteins of interest are expressed within the tissue and on what cell types. Moreover, in-depth quantitative data sets can be obtained when specific algorithms are applied to the rasters. Questions such as: (1) frequency of cells expressing marker(s) of interest; (2) cell type(s) in an area of interest; (3) expression level of markers of interest on certain cell types; and (4) relative distance of markers to the tumor or each other can be addressed. Thus, the data sets generated from MIBI-based analysis can be extremely large and in-depth to give a precise assessment of an individual or patient. Similarly, these types of data sets will be extremely informative for identifying patient populations that will respond to treatments during drug development.
Our current proof of concept study demonstrates that MIBI-analyzed samples can be used to quantitate protein expression level; however, there are limitations to the technology and our studies that restrict its use in practice. In terms of the technology, the field of view was only 100 by 100 μm, which is quite limited compared to the entire tissue section available to a pathologist. In addition, the collection time required to analyze each sample, which was ~27 min on the NanoSIMS instrument, is substantially longer compared to chromogen and AQUA image acquisition, which takes 1 min for each sample. These technical restrictions pose serious limitations to the high-throughput workflow required for tissue scoring. However, new instruments dedicated to imaging tissue sections can perform analysis faster by increasing sensitivity using a stronger source and time of flight analysis to reduce image acquisition time with increased field of view. In addition, the development of algorithms will permit faster data analysis through automation.
Another limitation for our studies is that the borders for the regions of interest were human drawn compared to an automated script that can identify cells and draw a uniform border around them. To account for that, cells that were not overlapping with other cells and entirely in the field of view were randomly chosen. In addition, borders were carefully drawn around the dsDNA/hematoxylin-merged rasters of the chosen cells without the HER2 raster to prevent any bias during demarcation. To show the mean pixel value analysis of these cells was representative of the sample, correlation analysis was also performed using total pixel count. A comparison of the correlation coefficients show the two parameters are comparable, suggesting that the mean pixel value analysis was representative of the image (Table 1).
Finally, MIBI analysis does not solve the pre-analytical factors that influence tissue section staining, such as variability in fixation/processing, storage and processing, epitope degradation, and poor antibody recognition.23 The FFPE samples destined for MIBI analysis come from standard sources and the staining process is similar to IHC protocols. In addition, the antibodies used for staining are the same clones and only differ with the metal tag for MIBI analysis. In this respect, MIBI is equivalent to IHC and cannot remedy the issues associated with pre-analytical factors. The difference with standard chromogenic IHC is the scoring, in which MIBI can be more objective and quantitative because subjective, semi-quantitative pathologist scoring is eliminated.
Despite these limitations and challenges, our studies show that quantitation of HER2 in breast carcinoma tissue from MIBI-analyzed samples strongly correlates with AQUA quantitation and pathologist-given IHC scores, demonstrating MIBI analysis as a viable method to measure protein expression. It is an initial step for validating a technology that has the ability to be objective and performs many of the features required for advancing diagnostics. It also has the power to answer questions in an in-depth manner that may introduce new avenues of thoughts. Taken together, MIBI analysis is a powerful technology that could be of great benefit for research, drug development, and treatment of patients.
References
Pegram MD, Pauletti G, Slamon DJ . HER-2/neu as a predictive marker of response to breast cancer therapy. Breast Cancer Res Treat 1998;52:65–77.
Hernandez-Blanquisett A, Touya D, Strasser-Weippl K et al, Current and emerging therapies of HER2-positive metastatic breast cancer. Breast 2016;29:170–177.
Vogel CL, Cobleigh MA, Tripathy D et al, Efficacy and safety of trastuzumab as a single agent in first-line treatment of HER2-overexpressing metastatic breast cancer. J Clin Oncol 2002;20:719–726.
Angelo M, Bendall SC, Finck R et al, Multiplexed ion beam imaging of human breast tumors. Nat Med 2014;20:436–442.
Levenson RM, Borowsky AD, Angelo M . Immunohistochemistry and mass spectrometry for highly multiplexed cellular molecular imaging. Lab Invest 2015;95:397–405.
Wolff AC, Hammond ME, Hicks DG et al, Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J Clin Oncol 2013;31:3997–4013.
Thomson TA, Hayes MM, Spinelli JJ et al, HER-2/neu in breast cancer: interobserver variability and performance of immunohistochemistry with 4 antibodies compared with fluorescent in situ hybridization. Mod Pathol 2001;14:1079–1086.
Koopman T, Louwen M, Hage M et al, Pathologic diagnostics of HER2 positivity in gastroesophageal adenocarcinoma. Am J Clin Pathol 2015;143:257–264.
Fox SB, Kumarasinghe MP, Armes JE et al, Gastric HER2 Testing Study (GaTHER): an evaluation of gastric/gastroesophageal junction cancer testing accuracy in Australia. Am J Surg Pathol 2012;36:577–582.
Ruschoff J, Dietel M, Baretton G et al, HER2 diagnostics in gastric cancer-guideline validation and development of standardized immunohistochemical testing. Virchows Arch 2010;457:299–307.
Warneke VS, Behrens HM, Boger C et al, Her2/neu testing in gastric cancer: evaluating the risk of sampling errors. Ann Oncol 2013;24:725–733.
Rimm DL . What brown cannot do for you. Nat Biotechnol 2006;24:914–916.
Dowsett M, Hanna WM, Kockx M et al, Standardization of HER2 testing: results of an international proficiency-testing ring study. Mod Pathol 2007;20:584–591.
Perez EA, Suman VJ, Davidson NE et al, HER2 testing by local, central, and reference laboratories in specimens from the North Central Cancer Treatment Group N9831 intergroup adjuvant trial. J Clin Oncol 2006;24:3032–3038.
Press MF, Sauter G, Bernstein L et al, Diagnostic evaluation of HER-2 as a molecular target: an assessment of accuracy and reproducibility of laboratory testing in large, prospective, randomized clinical trials. Clin Cancer Res 2005;11:6598–6607.
Umemura S, Osamura RY, Akiyama F et al, What causes discrepancies in HER2 testing for breast cancer? A Japanese ring study in conjunction with the global standard. Am J Clin Pathol 2008;130:883–891.
Illy N, Majonis D, Herrera I et al, Metal-chelating polymers by anionic ring-opening polymerization and their use in quantitative mass cytometry. Biomacromolecules 2012;13:2359–2369.
Lou X, Zhang G, Herrera I et al, Polymer-based elemental tags for sensitive bioassays. Angew Chem Int Ed Engl 2007;46:6111–6114.
Carvajal-Hausdorf DE, Schalper KA, Pusztai L et al, Measurement of domain-specific HER2 (ERBB2) expression may classify benefit from trastuzumab in breast cancer. J Natl Cancer Inst 2015;107:djv136.
Camp RL, Chung GG, Rimm DL . Automated subcellular localization and quantification of protein expression in tissue microarrays. Nat Med 2002;8:1323–1327.
Cayer DM, Nazor KL, Schork NJ . Mission critical: the need for proteomics in the era of next-generation sequencing and precision medicine. Hum Mol Genet 2016;25:R182–R189.
Giesen C, Wang HA, Schapiro D et al, Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat Methods 2014;11:417–422.
O'Hurley G, Sjostedt E, Rahman A et al, Garbage in, garbage out: a critical evaluation of strategies used for validation of immunohistochemical biomarkers. Mol Oncol 2014;8:783–798.
Acknowledgements
Part of this work was conducted using the CAMECA NanoSIMS 50 L, supported by the NSF under award # 0922648. We thank the Genentech Pathology Core group for help in procuring the breast carcinoma tissue, generating the TMAs, and imaging of chromogenically stained samples.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
SR, JG, HK, and SDL are employees of Genentech; JMB is an employee of Navigate BioPharma Services. The remaining author declares no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Laboratory Investigation website
Multiplexed ion-beam imaging analysis (MIBI) is a tissue section analysis technology that uses mass spectrometry to detect metal-tagged detection reagents. Quantitation based on MIBI analysis corresponds with pathologistdetermined and immunofluorescence-based automated quantitative analysis scores in a blinded study using the well-established HER2 model. These studies provide initial validation for quantitation of protein expression level by MIBI.
Supplementary information
Rights and permissions
About this article
Cite this article
Rost, S., Giltnane, J., Bordeaux, J. et al. Multiplexed ion beam imaging analysis for quantitation of protein expresssion in cancer tissue sections. Lab Invest 97, 992–1003 (2017). https://doi.org/10.1038/labinvest.2017.50
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/labinvest.2017.50
This article is cited by
-
Spatial analysis of the osteoarthritis microenvironment: techniques, insights, and applications
Bone Research (2024)
-
Mass spectrometry imaging in gynecological cancers: the best is yet to come
Cancer Cell International (2022)
-
Spatially resolved absolute quantitation in thin tissue by mass spectrometry
Analytical and Bioanalytical Chemistry (2021)
-
Multiplexed ion beam imaging (MIBI) for characterization of the tumor microenvironment across tumor types
Laboratory Investigation (2020)