Abstract
Analysis of specific DNA alterations in precision medicine of tumors is crucially important for molecular targeted treatments. Lung cancer is a prototypic example and one of the leading causes of cancer-related deaths worldwide. One major technical problem of detecting DNA alterations in tissue samples is cellular heterogeneity, that is, mixture of tumor and normal cells. Microdissection is an important tool to enrich tumor cells from heterogeneous tissue samples. However, conventional laser capture microdissection has several disadvantages like user-dependent selection of regions of interest (ROI), high costs for dissection systems and long processing times. ROI selection in expression-based microdissection (xMD) directly relies on cancer cell-specific immunostaining. Whole-slide irradiation leads to localized energy absorption at the sites of most intensive staining and melting of a membrane covering the slide, so that tumor cells can be isolated by removing the complete membrane. In this study, we optimized xMD of lung cancer tissue by enhancing staining intensity of tumor cell-specific immunostaining and processing of the stained samples. This optimized procedure did not alter DNA quality and resulted in enrichment of mutated EGFR DNA from lung adenocarcinoma specimens after xMD. We here also introduce a quality control protocol based on digital whole-slide scanning and image analysis before and after xMD to quantify selectivity and efficiency of the procedure. In summary, this study provides a workflow for xMD, adapted and tested for lung cancer tissue that can be used for lung tumor cell dissection before diagnostic or investigatory analyses.
Similar content being viewed by others
Main
Low tumor cell content is a major problem in the investigation of cancer cell-specific molecular mechanisms in patient derived tissue samples. Undissected samples contain varying proportions of non-neoplastic cells, including the tumor stroma, inflammatory infiltrate and adjacent normal tissue. Undesired normal DNA, RNA or proteins can dilute the tumor cell signatures so that important molecular alterations, such as activating mutations may fall below detection levels. One solution is the enrichment of the desired cell population before subsequent nucleic acid or protein extraction. Laser capture microdissection was invented in the 1990s and since then has evolved as a valuable tool for separating tumor cells from the surrounding tumor stroma.1, 2, 3, 4 However, conventional laser capture microdissection harbors several disadvantages including user dependent, subjective selection of cancer cells, high costs for commercially available dissection systems and because of the cell-by-cell selection procedure has very long processing times.
In 2011, Hanson et al5 described a workflow of user-independent expression-based microdissection (xMD) adapted to commercially available laser dissection instruments and also to a handheld laser device based on previous description of xMD from 2004 by Tangrea et al.6 This workflow provides a tool for fast microdissection of large amounts of tumor cells for further downstream analysis. In this workflow, tumor tissue is immunostained by antibodies against tumor-specific antigens and sites of immunoprecipitation are visualized with a dark chromogen such as diaminobenzidine (DAB). Subsequently, the stained slices are covered with an ethylene-vinyl-acetate (EVA) membrane. Laser irradiation of the complete slide leads to absorption and heating of the darkly stained tumor cells and to a localized melting of the EVA membrane, so that cell selection for microdissection is based on immunohistochemistry (IHC) rather than on user-dependent cell-by-cell collection. Afterward, the EVA membrane with the attached tumor cells or even subcellular compartments, for example, the isolated nucleus can be removed so that the tumor cells are separated from the tumor microenvironment.5, 6, 7 The success and specificity of the microdissection can be verified by comparing the tissue sections before and after the procedure.
Molecular diagnostics in lung cancer, one of the leading causes of cancer-related death worldwide,8 is particularly challenging as most patients are diagnosed in advanced tumor stages without the option for tumor resection. In these patients with an urgent need for molecular-guided medical treatments, small biopsies containing only few tumor cells and a majority of non-neoplastic cells are the only tissue available. Comprehensive genetic studies revealed multiple subgroups of lung cancer9, 10 and genetic alterations in genes encoding for protein kinases such as epidermal growth factor receptor (EGFR) or anaplastic lymphoma kinase are highly predictive for a durable clinical response to small molecule inhibitors,11, 12 if their mutation status is determined with high precision. Microdissection can enrich tumor cells in small biopsies in order to increase the molecular diagnostic sensitivity.
In this study, we demonstrate an optimized method to enrich lung adenocarcinoma tumor cells from heterogeneous tissue samples. First, we optimized the technique of xMD of lung cancer tissue with a handheld laser device. For this purpose, we optimized (1) chromogenic precipitation during IHC, (2) the treatment of the stained slices and (3) the handling of the laser. Second, we evaluated DNA quality derived from EVA membrane and showed enrichment of mutated EGFR DNA from lung cancer specimens after xMD. Third, we investigated microdissection selectivity and efficiency by pre-post whole-slide scanning and image analysis.
MATERIALS AND METHODS
Ethics Statement
Tissue samples of lung cancer patients were obtained from surgical lung resections at the thoracic surgery department of the University Medical Center Göttingen. All patients gave informed consent to participate in this study. All procedures were conducted in accordance with the Declaration of Helsinki and institutional, state and federal guidelines. The use of lung cancer specimens for this study was approved by the ethics committee of the University Medical Center Göttingen (approval #1-2-08).
Immunostainings of Lung Cancer Samples
Lung adenocarcinoma tissue samples were acquired from patients who underwent lobectomy surgery at the Department of Thoracic and Cardiovascular Surgery in the period from 2012 to 2015. The samples were fixed immediately after resection in 4% neutral-buffered formalin overnight and embedded in paraffin. Immunohistochemical reactions were performed on 10 μm sections. After deparaffinization and rehydration with decreasing concentrations of ethanol, antigen retrieval was performed by incubation in citrate buffer (pH 6.1) at 90 °C for 40 min. Anti-CK7 primary antibody OV-TL 12/30 (Dako, Glostrop, Denmark) was incubated for 60 min at room temperature at a concentration of 1:50. Thereafter, the sections were incubated for 30 min with secondary antibodies coupled to biotin and subsequently 30 min with HRPO peroxidase coupled to streptavidin (REAL Detection System, K5005, Dako). DAB (Dako) or Vektor Black (Vector laboratories, Burlingame, CA, USA) was used as chromogen either following the instructions of the supplier or using excessive chromogen (3 × concentration). Hematoxylin counterstaining was not performed. Slides were dehydrated with ethanol and xylene, dried and left uncovered for processing.
Processing Steps for xMD
Immunostained lung cancer tissue sections were placed on a heating plate set to 78 °C and were covered by a section of EVA membrane with 19% vinyl-acetate content and 76 μm nominal thickness (CoTran 9715, 3M, St Paul, MN, USA). A sealing roller was applied at the beginning and after 5 min of heating to ensure uniform contact of the EVA membrane to the tissue section. A handheld laser device (Type SDL-15, Biotechnique Avance, UK) was used for the experiments and was set to the number two position for pulse frequency and to the number three setting for laser intensity. For laser safety precautions, all users wore laser safety goggles, lab coats and latex gloves. The stained tissue sample was irradiated with ~1500 laser pulses at an angle of 45° and a distance of ~1 cm. During exposure, visual feedback for successful impregnation of the EVA polymer was given by a wet appearance of the polymer caused by a reduction of diffuse light scattering, which in the first place results from a rough surface of the raw foil.
Selection and Characterization of the Radiation Source
The handheld laser device was characterized with regard to laser beam shape, power and pulse properties. Using a laser beam profiler (LaserCam-HR, Coherent Inc., CA, USA), the shape of the laser beam was recorded with a resolution of 6.7 μm and because of mechanical constraints (CMOS chip housing hitting the laser) at a distance of 48.1 mm from the laser outlet. In total, three images were captured along the line-shaped laser spot and stitched together using a self-written cross-correlation algorithm. Quantitative information was derived by normalization of the sum of pixel counts to the pulse energy.
The focal point was determined with help of a wedge laminated with stained paper: although lowering the wedge height using a micrometer screw, the paper was irradiated with single pulses. After the procedure, the focal point could be reliably read out from the intensity of the burn marks on the paper.
Power measurements were conducted using a calibrated thermopile sensor (PM10 measurement head, Coherent Inc.) at a distance of 35 mm from the laser outlet, enabling measurement of the whole beam. Owing to the low absorption of air, the distance does not affect the power readings unless the laser beam is clipped by the active area of the measurement head if the distance is too high. The value was determined after 3 s of exposure. Time resolved measurements of the pulses were accomplished using a combination of photodiode, preamplifier and transient recorder (M2i.3021, Spectrum GmbH, Germany). After characterization, the laser system was ranked among the other devices regarding the pulse fluence. For beam dimensions, the full width at half maximum (FWHM) was used. For complex beam profiles, a nonlinear least squares fit (using the Levenberg-Marquardt algorithm)13 to a Gaussian distribution was conducted before determining the FWHM.14 All computations were done using R (www.r-project.org).
DNA Isolation from EVA Membrane and Analysis of DNA Quality
DNA was extracted from the cells attached to the EVA membrane, the remnant cells and an untreated control slide using the GeneRead DNA FFPE Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol, but with the following modification: skipping of the deparaffinization step, doubling of the proteinase K enzyme solution and extension of the digestion to 2 h. Concentration of the extracted DNA was determined using a droplet digital PCR Taqman assay targeting a 91-bp region on chromosome 5:13702190–13702281. The amplified copies/μl DNA solution as obtained from digital droplet PCR were multiplied by 3.3 pg (weight of haploid human genome) and by the total extraction volume in order to calculate the amount of amplifiable DNA in ng/μl. Primer sequences were: FWD: 5′-TGCTTCTGACTTTCTCCCTGA-3′ and REV: 5′-ACACTGCTGCTGCTGATGAC-3′, the internal probe was labeled with HEX and quenched by Blackhole Quencher 1 (BHQ1). Reactions were conducted in a total reaction volume of 20 μl in 1 × ddPCR Super-Mix for Probes (Bio-Rad, Hercules, USA) with 250 nM of probe and 900 nM of each primer. The EGFR E746-A750del mutation and the corresponding wild-type allele were detected using allele-specific primers (EGFRdelA746-A750.FWD: 5′-AATTCCCGTCGCTATCAAAAC-3′; EGFR_wildtype.FWD: 5′-CCGTCGCTATCAAGGAATTAAG-3′), a universal reverse primer (EGFR.REV: 5′-CCACACAGCAAAGCAGAAA-3′) and a universal probe (EGFR.Probe: FAM-5′-CCAACAAGGAAATCCTCGATGT-3′-BHQ1). Each template was tested in two ddPCRs (EGFRdelA746-A750 and wild-type), which were loaded with the same amount of template (20 ng). Reactions were conducted as described above with primer concentrations lowered to 225 nM of each primer.
Slide Scan and Image Analysis
Whole-slide scanning was performed using a Pannoramic P250 slide-scanner (3DHistech, Budapest, Hungary) at a resolution of 4.5 μm/pixel. Image analysis was performed using ImageJ version 1.48 (National Institutes of Health, USA; http://imagej.nih.gov/ij). Pre- and post-processing images were aligned and differences in intensity were calculated. For visualization, the preprocessing images were merged with pseudo-color overlays of the intensity differences that represent the decrease in staining intensity, that is, the transferred tissue. Scatter plots of the intensity differences were calculated with 'R' version 3.2.2 (www.r-project.org). Staining intensities were represented as 8 bit gray-scale values (28=0–255; 0≙white, 255≙black) or as relative staining intensity (0–1).
RESULTS
Optimization of the Preprocessing of Lung Cancer Tissue Samples for xMD
xMD relies upon effective and specific immunohistochemical staining of the tumor cells, because absorption of the near infrared laser energy depends largely on the staining darkness, which results in localized heating and melting of the EVA membrane that covers the stained tissue. In order to optimize the staining darkness, we tested different chromogens in combination with an anti-CK7 primary antibody on formalin-fixed lung adenocarcinoma tissue samples and found Vektor black (Vektor Laboratories) leading to a dark black staining of tumor cells, especially if excess substrate was applied by increasing the concentration threefold (Figures 1a–d).
Optimized IHC and xMD procedure. Tissue sections of lung adenocarcinoma samples were stained with hematoxylin and eosin (a) or immunohistochemically stained with anti-CK7 antibodies visualized by DAB (b) or Vektor Black following the instructions of the supplier (c) or using excessive chromogen (3 × concentration of Vektor Black) (d). A 40-fold magnification is shown (a-d). For xMD, immunostained lung cancer tissue sections were placed on a heating plate and a handheld laser device was used for the irradiation of the slide in an angle of 45° (e).
To further optimize the localized melting of the EVA membrane by lowering the necessary energy from the laser source, we applied constant heating of the stained tissue slide by a heating plate, which increases microdissection efficiency (Figure 1e). A temperature of 78 °C was established to be optimal for lung cancer microdissection.
Finally, we tested the effect of tissue section thickness on the microdissection efficacy and found a section thickness of 10 μm is optimal. Thinner tissue sections (< 10 μm) showed a reduced detachment of cells from the glass slide to the EVA membrane. Thicker sections (> 10 μm) showed a complete detachment during sample preparation and can lead to cell overlay.
Analysis of Radiation Source
Besides suitable preprocessing steps, the right choice of both radiation source and exposure parameters are crucial for successful xMD.1, 15 Four different laser sources have been tested by Hanson et al:5 three laser capture microdissection microscopes and a consumer class depilatory laser. The latter (SDL-15) was chosen for our experiments as it is easily available, as well as operator friendly and features a line-focused beam, which is well suited for wide-area exposure.
The SDL-15 device encompasses a gallium arsenide laser diode with a nominal emission wavelength of 808 nm. The diode is mounted on an aluminum block. Thermal management is supported by a small fan attached to the back of the aluminum block. The laser line spot is formed by a cylindrical lens focusing the laser emission along one direction (causing the width of the line spot) although leaving the light propagation in the perpendicular direction unchanged (causing the length of the line spot). The pulses are generated by modulation of the pumping current. Two parameters are offered by the stationary laser driver: laser power and pulse repetition rate, each adjustable to three values. Nominally, the repetition rates are 3, 5 and 7 pulses per second. The measured values, on the other hand, were 7.1, 8.8 and 9.5 per second. For all three levels, the pulse duration was 33.7 ms.
The nominal optical output power (‘total laser power’) is 15 W. Power measurements yielded an average power of 547 and 2459 mW for the second and third position of the intensity knob, respectively, while using the highest repetition rate. The resulting continuous wave powers are 1.7 and 7.7 W, respectively, which is approximately half of the nominal value of 15 W. When using the highest intensity, a power reduction of 3% after 20 s was observed. After some usage with regular breaks in between, a power drop of up to 10% was observed. After a break of approximately 6 min, the original power was restored. With the intensity at level two, the maximum power loss was only 3%. Here, the laser fully recovered after a pause of only 60 s.
The focal point was determined at 17.75 mm distance from the laser outlet. The length of the line-shaped laser beam was 5.87 mm and the line width in the fast axis was about 108 μm within this distance (Figure 2). Accordingly, this resulted in an average per pulse fluence of 41 J/cm2 (range 24–59 J/cm2) in the focal plane.
Profile of the laser beam. Using a laser beam profiler, the length and the width of the laser beam in a distance of 48.1 mm from the laser diodes outlet was recorded with a resolution of 6.7 μm. Three images were captured and stitched together, quantitative information are shown by normalization the sum of pixel counts to the pulse energy.
Hanson et al5 tested four different laser systems and recommended sets of operation parameters for each system. Table 1 summarizes the results and also shows derived and measured parameters. The pulse energy was calculated under the assumption that the continuous wave and not the average powers of the lasers were given.
Compared with the pulse fluences of other systems, the SDL-15 system is rather in the lower range of the values reported as working for xMD (Table 1), especially when attributing the low fluence of the Veritas system to the low pulse duration, which compensates for heat dissipation. When considering the original recommendation of 2–3 cm working distance and an angle of 45° (the angle causes a sin(α) factor), the fluence of the SDL-15 lies between 9.7 and 26 J/cm2, which is very low. In order to maximize the fluence while maintaining spot area and operability, a distance of about 1 cm and an angle of 45° were chosen for our experiments. In this case, the spacer bar in front of the laser outlet can be used as support. Furthermore, a power setting of level three and a frequency setting of level two was chosen for our experiments. In this configuration, a per pulse fluence of 51 J/cm2 and an approximate spot area of 0.5 mm2 can be expected. With an assumed spot area overlap of 50% as reported in the original protocol,5 a quadratic tissue area with an edge length of about 19.4 mm can be processed when applying a total number of 1500 pulses (equals about 2.84 min without breaks). This calculation corresponds well with the observations we made during xMD processing.
For thermal control, the laser was used at frequency level 2 for 60 s, alternating with 60 s of break for a maximum time of 10 min before making a 5-min break.
Enrichment of Mutated DNA from Lung Cancer Specimen after Microdissection
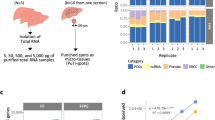
Mutational analysis of lung cancer tissue can be challenging, if only few tumor cells are surrounded by an extensive tumor stroma, so that microdissection technologies are helpful to enhance sensitivity. In particular, as DNA extracted from FFPE specimen is often highly fragmented and/or chemically modified,16 which contributes to loss in sensitivity. In order to test if IHC or subsequent xMD alters the quality of the DNA isolated from EVA membranes, the integrity was first assessed by capillary electrophoresis of DNA isolated from one EVA membrane after xMD (Figure 3a), which yielded a median fragment length of approximately 650 bp, which is comparable to DNA isolated from undissected FFPE tissue. In addition, DNA was isolated from unstained and undissected samples (control), from EVA membrane after dissection (EVA) and also from the not-transferred remaining tissue on the glass slide after microdissection (remnant) from four different lung adenocarcinoma samples. The amount of DNA that was amplifiable by a 91-bp amplicon was quantified by droplet digital PCR (Figure 3b). 1D plots of the droplet digital PCR of these four samples are shown in Figure 3c. Optimally, two distinct bands of negative (black) and positive (green) are seen. However, especially when analyzing fragmented DNA such as FFPE DNA, droplets with lower fluorescence amplitudes are frequently seen. Such droplets are commonly termed as ‘rain’ and presumably contain chemically modified and/or fragmented template molecules that negatively impact amplification efficiency during PCR. Comparing the DNA extracted from the control slices with the EVA membrane and remnant DNA the degree of ‘rainfall’ was not increased, suggesting no additional DNA damage is caused by IHC and microdissection.
DNA analysis of microdissected lung cancer tissue. Electropherogram of DNA isolated from EVA membrane after xMD showing a median fragment length of approximately 650 bp (a). Amount of amplifiable DNA isolated from EVA membrane after xMD derived from four different lung cancer samples tested by a 91-bp amplicon and quantified by droplet digital PCR (b). 1D plots of the droplet digital PCR of the 91-bp amplicon, showing the green-labeled positive and the black-labeled negative band of amplified DNA (c). The ratio of mutated EGFR (E746-A750del) to wild-type EGFR DNA isolated from unstained and undissected samples (control), from EVA membrane after dissection (EVA) and from the untransferred remaining tissue on the glass slide after microdissection (remnant) from three different lung adenocarcinoma tissue samples with known EGFR mutation as obtained by digital droplet PCR (d).
Furthermore, three additional lung adenocarcinoma tissue samples with known EGFR mutation (E746-A750del) were used to quantify the ratio of mutated EGFR compared with wild-type EGFR by droplet digital PCR. When DNA isolated from unstained and undissected samples (control), from EVA membrane after dissection (EVA) and from the not-transferred remaining tissue on the glass slide after microdissection (remnant) was compared, an increase of the mutated to wild-type EGFR ratio after microdissection was demonstrated, whereas the DNA isolated from the remnant showed a depletion of mutated EGFR (Figure 3d).
Comparative Image Analysis of Tissue Samples before and after Microdissection
Successful melting of the EVA polymer can be directly observed as the polymer gets a 'wet' appearance when sufficiently heated. For verification of the xMD-transfer, the IHC-stained, uncovered tissue sections can be checked by light microscopy before and after the procedure.
To document and quantify the pre- and post-transfer changes, we performed whole-slide scanning and image analysis. Figure 4 illustrates one resection specimen, that is, the tissue before microdissection (Figure 4a1; enlarged detail: Figure 4b1), a visualization of decreased staining intensity corresponding to the transferred tissue (Figures 4a2 and b2) and the tissue after microdissection (Figure 4a3 and b3). Histograms of the subregions show that the intensity decreased in the transferred areas, whereas the untransferred regions showed no change in intensity (Figure 4c). For quantification, the intensities of each corresponding pixel before and after xMD were plotted (Figure 4d1). The resulting scatter plot shows that areas with high staining intensity (>100) on the pre-treatment scan show reduced staining intensity on the post-treatment scan (0–100). Areas with moderate or low staining intensity show comparable values on both scans. The effect is easier to demonstrate if the median value is calculated for each staining intensity (0–255, Figure 4d2). Thus, it seems that the transfer affected mostly the dark stained tumor cells, whereas the light stained non-neoplastic cells were preferentially omitted.
Image analysis of microdissected lung cancer tissue. One cytokeratin 7 immunostained lung cancer specimen is shown before (a1; enlarged detail: b1) and after xMD (a3; enlarged detail: b3). Areas of decreased staining intensity corresponding to the transferred regions are visualized by pseudo colors (a2, enlarged detail: b2). Histograms of the transferred and untransferred regions illustrate the decrease in staining intensity (c). For quantification, the intensity before and after transfer of each corresponding pixel are plotted (d1) and the median value is calculated for each staining intensity (d2).
As negative control, we performed a mock treatment of two slides by omitting the laser irradiation (Figures 5a and b,Supplementary Figures 2A and B). As expected, comparisons of whole-slide scans before and after mock treatment showed no difference in staining intensity: although the scatter plot showed some changes, the medians formed a line that was almost in perfect agreement with the diagonal line. Thus, the scatter plot appeared to show random noise caused by the repeated scanning and image alignment; any direct effect would have caused deviation of the medians from the diagonal.
Image analysis was verified by making artificial modifications to the preprocessing images (Supplementary Figure 1): no changes to the preprocessing image resulted in a diagonal line for both plots (Supplementary Figure 1A), increasing the brightness shifted the interception of the diagonal (Supplementary Figure 1B) and increasing the contrast shifted the slope (Supplementary Figure 1C). Adding random noise caused the 2D plot to form a dense point cloud around the original diagonal, whereas the median values showed a near-perfect line (Supplementary Figure 1D). Thus, plotting the medians per intensity category seems to be a suitable visualization to exclude random effects and show true, directed intensity changes.
Finally, reproducibility of our optimized xMD workflow for lung cancer tissue was demonstrated by a series of nine independently stained lung cancer sections (Figures 5c–f,Supplementary Figures 2C–K).
DISCUSSION
Analysis of specific DNA alterations in precision oncology is crucially important for molecular targeted treatments. Lung cancer is a prototypic example and one of the leading causes of cancer-related deaths worldwide. Diagnostics on FFPE biopsies were shown to detect a specific EGFR (T790M) mutation with a sensitivity of only 70%,17 clearly illustrating the need for further improvement. It is likely that a combination of both, plasma-derived and enriched FFPE DNA mutation testing, would allow to further maximize the number of cancer patients that can benefit from targeted therapies. One major reason for the suboptimal sensitivity in tissues is the fact that especially lung cancer samples are often limited to small biopsies consisting of a heterogeneous mixture of cancer cells, their surrounding stroma and immune cells. Therefore, many diagnostic analyses, in particular the detection of genetic alterations, require tiresome microdissection techniques before DNA isolation in order to increase the sensitivity and specificity of the analysis and to ensure correct results. xMD has several advantages in comparison with conventional laser capture microdissection, as the sample handling time is shorter and as selection of cells for microdissection relies on protein expression and antibody stainings rather than ‘subjective’ user-dependent choice. However, it has been shown in several contexts that efficiency and specificity of xMD relies strongly on immunostaining intensity and the applied laser energy so that both IHC and laser irradiation must be adapted for every new tissue context.18, 19, 20 Furthermore, also downstream analyses like DNA-based methods will potentially be influenced by sample handling and should be evaluated after microdissection in comparison with undissected tissue samples.21
Therefore, in the first step of this study, we optimized xMD for pulmonary adenocarcinomas. For this purpose, we enhanced immunohistochemical chromogenic precipitation and thus staining intensity by using an anti-cytokeratin-7 primary antibody and Vektor Black as chromogen leading to a dark black staining that absorbs energy of the infrared laser irradiation better than tissue slides stained with DAB. CK7 was used because it allows for an intense immunohistochemical staining of lung adenocarcinoma cells. However, it is reasonable to assume that Vektor Black will also enhance dissection efficiency in the context of other tissue samples and immunostainings. xMD is certainly not limited to cytokeratines and should work for every immunostaining that can be stained intensively, also for example, for single immune cells, but a thorough optimization of the staining intensity should be performed for every new setting.
Furthermore, we found that heating of the tissue samples covered with the EVA membrane can strongly increase dissection efficiency. The EVA membrane used in this study (CoTran 9715, 3 M) contains 19% of vinyl-acetate and has a thickness of 76.2 μm. The melting point of this membrane is approximately 80 °C and the preheating lowers the energy that is needed from the laser device to melt the EVA membrane. It is important to notice that the correct adjustment of the temperature is crucial, as low temperatures will decrease dissection efficiency and high temperatures will lead to unspecific melting of the membrane. In our study, we found a temperature of 78 °C to be optimal for dissection efficiency, but the temperature can vary using different tissue samples, different EVA membranes or different laser devices and needs to be optimized for every new setting. However, as in the case of Vektor Black, the preheating of a tissue sample before laser irradiation could enhance dissection efficiency also for various other tissue specimens.
By a technical analysis of the handheld laser device, which is both easy to handle and inexpensive compared with classical LCM-devises, we could develop a set of recommendations for optimized laser handling. The per pulse fluence (energy per area per pulse) was used as an important indicator for the analysis and comparison of different devices used for xMD. The fluence is often used to define thresholds of effect (eg, for ablation of material). With the pulse durations being in the range of milliseconds, however, heat conduction is not negligible and the pulse duration has to be considered as well (see deviations of the Veritas LCM device). In general, the parameters found in the literature and the ones that were measured and tested here were consistent when regarding the per pulse fluence. However, the required information about laser power and spot diameter strongly depend on definitions (eg, FWHM or 1/e2 spot boundaries, power before or after the optics). Thus, a comprehensive parameter study under uniform measurement conditions aiming at the specific challenges of xMD (eg, selectivity and efficiency) should be conducted. Although a line-shaped spot of the SDL-15 system is a first step toward area exposure, a homogenization of the laser beam would further improve and simplify the exposure. Finally, parameters like laser power, working distance or spot overlap should be invariant during the process in order to ensure maximum repeatability.
Subsequently, we evaluated DNA quality derived from EVA membranes in order to test, if immunostaining sample handling or laser irradiation alters the DNA integrity. For this purpose, we evaluated DNA-fragment length isolated from EVA membranes after xMD and found a median fragment length of nearly 650-bp that is comparable to DNA isolated from FFPE tissue samples without any further dissection. The DNA amount that could be isolated from EVA membranes was lower than the amount derived from undissected controls, which is reasonable as tumor stroma and immune cells were excluded by microdissection, However, the lower amount of isolated DNA was sufficient for amplification and quantification of an 91-bp amplicon by droplet digital PCR. In comparison with unstained FFPE samples to the stained remnant that was not dissected after xMD and to DNA isolated from EVA membrane, there were no relevant differences in the degree of droplets with lower fluorescence (‘rainfall phenomenon’) suggesting that DNA quality was not affected either by immunostaining or by laser irradiation or the EVA membrane. As shown here, the method was able to enrich EGFR-mutated DNA from lung adenocarcinoma tissue specimens. As activating EGFR mutations affect only one allele of the DNA, a pure population of EGFR-mutated cells would show a ratio (mutated/unmutated) of 1. xMD enriched the mutation positive cells to 89% of the maximal achievable enrichment for samples that were at approximately 30% in the original tissue; in one samples with 70% original content the enrichment was 93% of maximum. We therefore conclude that DNA isolated from EVA membranes after xMD is suitable for further diagnostic and scientific procedures.
Finally, we used whole-slide scanning before and after xMD and digital image analysis for a comprehensive evaluation of microdissection selectivity and efficiency. Successful microdissection can be assessed on different steps of the procedure. Successful melting of the EVA membrane can be observed, when a wet appearance of the polymer becomes visible. Furthermore, assessment of slides and the EVA membrane before and after the process can be done visually by microscopic estimation. However, whole-slide scanning and digital image analysis provide a complete and accurate analysis of efficiency and specificity of xMD of lung adenocarcinoma, as a reduction of staining intensity can be detected quantitatively for every single pixel.
Visualization by histograms showed a cutoff in staining intensity, which divided the low staining areas (ie, areas with predominance of normal cells) from the high staining areas (ie, areas enriched for tumor cells). Mainly the areas of high staining were depleted after xMD. The reproducibility of selective and efficient microdissection by xMD was shown on nine separate stainings. However, in some of the samples we also observed a low, but notable decrease of intensity in the low intensity areas because of unspecific microdissection, and some areas of high staining intensity without reduction of intensity after the process. Thus, we show that xMD is a very promising technology that enables a selective, efficient and reproducible microdissection. In our view, the main component that will require further improvement is the laser source. One possibility is to use flash lamps instead of laser devices,7, 20 another one could be to test different laser sources like stationary systems (eg, Arcturus) with higher and more reproducible fluence. If markers other than CK7 should be used, the immunostaining must also be carefully optimized.
In conclusion, we here provide a rapid and reproducible microdissection workflow that was evaluated for subsequent DNA-based mutational analyses in lung adenocarcinomas. We believe that this workflow can be used for many diagnostic and scientific purposes and will serve as a basis for further protocol development.
References
Emmert-Buck MR, Bonner RF, Smith PD et al, Laser capture microdissection. Science 1996;274:998–1001.
Bonner RF, Emmert-Buck M, Cole K et al, Laser capture microdissection: molecular analysis of tissue. Science 1997;278:1481–1483.
Hunt JL, Finkelstein SD . Microdissection techniques for molecular testing in surgical pathology. Arch Pathol Lab Med 2004;128:1372–1378.
Espina V, Wulfkuhle JD, Calvert VS et al, Laser-capture microdissection. Nat Protoc 2006;1:586–603.
Hanson JC, Tangrea MA, Kim S et al, Expression based microdissection adapted to commercial laser dissection instruments. Nat Protoc 2011;6:457–467.
Tangrea MA, Chuaqui RF, Gillespie JW et al, Expression microdissection: operator-independent retrieval of cells for molecular profiling. Diagn Mol Pathol 2004;13:207–212.
Blackler AR, Morgan NY, Gao B et al, Proteomic analysis of nuclei dissected from fixed rat brain tissue using expression microdissection. Anal Chem 2013;85:7139–7145.
World Health Organization. GLOBOCAN 2008 - Estimated cancer Incidence, Mortality, Prevalence and Disability-adjusted life years (DALYs) Worldwide in 2008. Available from: URL http://globocan.iarc.fr/.
The Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012;489:519–525.
The Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511:543–550.
Lynch TJ, Bell DW, Sordella R et al, Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 2004;20:2129–2139.
Soda M, Choi YL, Enomoto M et al, Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 2007;448:561–566.
Moré JJ . The Levenberg-Marquardt algorithm: implementation and theory. Numerical Anal 1978;630:105–116.
Liu JM . Simple technique for measurements of pulsed Gaussian-beam spot sizes. Optics Lett 1982;7.5:196–198.
Goldstein SR, McQueen PG, Bonner RF . Thermal modeling of laser capture microdissection. Appl Optics 1998;37:7378–7391.
Didelot A, Kotsopoulos SK, Lupo A et al, Multiplex picoliter-droplet digital PCR for quantitative assessment of DNA integrity in clinical samples. Clin Chem 2013;59:815–823.
Oxnard GR, Thress KS, Alden RS et al, Association between plasma genotyping and outcomes of treatment with osimertinib (AZD9291) in advanced non-small-cell lung cancer. J Clin Oncol 2016;34:3375–3382.
Grover AC, Tangrea MA, Woodson KG et al, Tumor-associated endothelial cells display GSTP1 and RARbeta2 promoter methylation in human prostate cancer. J Transl Med 2006;4:13.
Hanson JA, Gillespie JW, Grover A et al, Gene promoter methylation in prostate tumor-associated stromal cells. J Natl Cancer Inst 2006;98:255–261.
Du Q, Yan W, Burton VH et al, Validation of esophageal squamous cell carcinoma candidate genes from high-throughput transcriptomic studies. Am J Cancer Res 2013;3:402–410.
Tangrea MA, Mukherjee S, Gao B et al, Effect of immunohistochemistry on molecular analysis of tissue samples: implications for microdissection technologies. J Histochem Cytochem 2011;59:591–600.
Acknowledgements
We thank Jennifer Appelhans and Mercedes Martin-Ortega for their technical support. Hanibal Bohnenberger was supported by the Else-Kröner-Fresenius-Foundation. We acknowledge support by the German Research Foundation and the Open Access Publication Funds of the Göttingen University.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Laboratory Investigation website
This study provides a workflow for expression-based microdissection adapted for lung adenocarcinoma. The authors improved microdissection by optimizing immunostaining intensity, processing of the stained samples and by providing instructions for laser handling. Using this procedure, the authors could enrich mutated EGFR-DNA from lung adenocarcinoma tissue samples. Selectivity and efficiency of microdissection was further demonstrated by pre-post whole-slide image analysis.
Supplementary information
Rights and permissions
About this article
Cite this article
Grafen, M., Hofmann, T., Scheel, A. et al. Optimized expression-based microdissection of formalin-fixed lung cancer tissue. Lab Invest 97, 863–872 (2017). https://doi.org/10.1038/labinvest.2017.31
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/labinvest.2017.31