Abstract
In cancer progression, metastasis is a major cause of poor survival of patients and can be targeted for therapeutic interventions. The first discovered metastatic-suppressor Nm23-H1 possesses nucleoside diphosphate kinase, histidine kinase, and DNase activity as a broad-spectrum enzyme. Recent advances in cancer metastasis have opened new ways for the development of therapeutic molecular approaches. In this review, we provide a summary of the current understanding of Nm23/NDPKs in the context of viral oncogenesis. We also focused on Nm23-H1-mediated cellular events with an emphasis on chromatin modifications. How Nm23-H1 modulates the activities of chromatin modifiers through interaction with Epstein–Barr virus-encoded oncogenic antigens and related crosstalks are discussed in the context of other oncogenic viruses. We also described the current understanding of the cellular and viral interactions of Nm23-H1 and their reference to transcription regulation and metastasis. Further, we summarized the recent therapeutic approaches targeting Nm23 and its potential links to pathways that can be exploited by oncogenic viruses.
Similar content being viewed by others
Introduction
Oncogenesis is more than a proliferating aggregated mass of cells. In the progression to the cancer phenotype, normal cells acquire signature attributes that transform them into tumorigenic and eventually malignant cells with metastatic potential. Metastasis is a phenomenon where cells with tumorigenic potential evade the nearby tissues and spread to distant organs from the primary neoplastic site.1, 2 This adaptation to a new microenvironment at distant tissues by primary tumor cells require expression of numerous cellular genes to reprogram cells and so bypass the evolutionary selection pressures.3 Furthermore, similar to other evolutionary events, a rare cell among millions wins this competition for survival.
More than 20 metastatic suppressors have been identified that influence many cellular signaling pathways and show metastatic-suppressor activities at different stages of metastasis, including intravasation, survival in the blood stream, extravasation, colonization and angiogenesis.4 Nm23-H1, the human isoform of Nme1 with NDPK activity, also called NDPK A (nucleoside diphosphatase kinase A), was the first metastatic-suppressor gene identified, and owing to an overwhelming abundance of experimental evidence, its metastatic-suppressor activity has been accepted beyond a doubt.5 In humans, the Nm23 family has 10 members (H1–H10) identified to date. Nm23-H1 and Nm23-H2 share 88% identity6, 7 but the antimetastatic activity of Nm23-H2 has not been fully explored.
As an essential function, NDPKs catalyze the exchange of phosphate groups between different nucleoside diphosphates and triphosphates and so sets up an equilibrium between different nucleoside triphosphates.8
The idea to look for tumor suppressors arose from the discovery of oncogenes. Bishop and Varmus investigated the concept that oncogenes influenced the growth and division of cells using the Rous sarcoma virus.9 They were the first to propose the idea that viral oncogenes are of cellular origin, hijacked during infection by viruses over time.9 The Nobel prize in Physiology and Medicine was awarded in 1989 for their discovery.9 This led to an explosion in the field with the discovery of cellular proto-oncogenes and tumor-suppressor genes.
Cancer cells have the attribute of genome-wide alterations, which results in dysregulation of the expression of cellular genes, and chromatin regulators important for normal regulation of cellular gene transcription. In cancer, various factors, including mutations in the regulatory genes, and epigenetic silencing can perturb the normal functioning of chromatin regulators, thus supporting the oncogenic behavior of cells.10 Chromatin regulators are responsible for packaging and unpacking DNA as well as modifying histones, thus regulating transcription, replication and repair of DNA.11 These chromatin regulatory proteins can be broadly classified into two groups either as chromatin modifiers, performing acetylation, methylation, ADP-ribosylation, ubiquitination and sumoylation of histone,11 or chromatin remodelers altering the structural confirmation of DNA–histone complex.11 Modifiers perform covalent modifications in an energy-independent manner, whereas remodelers utilize the ATP energy currency involved in packaging of DNA into nucleosome or unpacking through ejecting, translocating or inserting the histone octamer.12, 13
Human chromatin remodelers are organized into four groups of proteins. They include the SWI/SNF (SWItching defective/Sucrose Non Fermenting), ISW1 (Imitation SWItch), CHD (Chromodomain, Helicase, DNA binding) and INO80 (INOsitol requiring 80) families.11
Among chromatin modifiers, histone acetyl transferases (HATs) and histone deacetylases (HDACs) have contrasting actions.11 HATs manipulate the electrical status of the chromatin DNA complexes where acetylation removes the positive charge on the histone at various lysine residues reducing its binding capacity to negatively charged sugar-phosphate backbone of DNA and therefore access to transcription factors. HDACs perform the opposite activity in removing the acetyl groups on lysine residues on histones.14 There are 5 HAT families with 13 members, and 4 HDACs families having HDAC 1–11.11 Sirtuins (1–7) functions in many cellular processes, including inflammation, gene regulation, cell death and aging, through a range of activities. These include deacetylation, ADP-ribosyl-transferase, demyristoylation and desuccinylation.15, 16
Similarly, another contrasting functional pair of histone methyl transferase and histone demethylase add and remove methyl groups at lysine and arginine residues at different positions.17, 18 Histone methylation has varied effects on cellular functions as it interferes with the binding of transcription factors and other chromatin-binding proteins and performs the transcriptional activation/repression.11 Protein lysine methyl transferases have >50 members classified into 8 different groups and 9 protein arginine methyl transferases classified into 3 groups.11 These all can contribute to regulation of gene expression and may be targeted for dysregulation by oncogenic viral antigens.
ONCOGENIC PHENOTYPE INDUCED BY INFECTIOUS AGENTS
The carcinogenic process is a highly integrated series of complex events that leads to cell transformation and proliferation. Non-infectious carcinogens, viral carcinogens and other infectious agents capable of driving oncogenic phenotype are likely not the sole contributors to cancer. It is rather the accumulation of a series of events driven by interaction of carcinogens, infectious or non-infectious agents with host factors. These interactions can silence or suppress the cellular regulatory mechanisms, which protect cells against uncontrolled growth and proliferation.
Importantly, we should also mention that some common cancers are associated with non-infectious risk factors, which include age, alcoholism, exposure to chemical carcinogens, environmental pollution, chronic gastric inflammation, dietary habits, hormones, immunosuppression, obesity, exposure to radiation, sunlight and tobacco usage.19 These can further affect the susceptibility to other infectious agents as well as the microbiome, which impacts the homeostatic functions of many cellular processes and so causes uncontrolled growth and proliferation.
Oncogenic infectious agents, including viruses, are among the major contributors to human cancers. They contribute to approximately 20% of the cases.20 Epstein–Barr virus (EBV), Kaposi’s sarcoma associated herpes virus (KSHV) or human herpes virus 8, human papilloma virus (HPV), human T-lymphotropic virus-1 (HTLV-1), hepatitis B virus, hepatitis C virus (HCV) and Merkel cell polyomavirus (MCV) are viruses well known to directly contribute to the development of different cancers (Figure 1).
A schematic showing the different tumor viruses, their tumorigenic determinants and associated diseases. Tumorigenic viruses such as EBV, KSHV, HPV, etc infect the human host, and through the direct and indirect interactions, viral antigenic determinants dysregulate the protective signaling mechanisms against growth of cancer cells. Examples of well-studied viral antigenic determinants are EBNA3C of EBV and LANA of KSHV.
Several viral factors such as LMP1 and EBNA3C of EBV; E6 and E7 of HPV; latency-associated nuclear antigen (LANA), vIRF and vCyclin of KSHV, large and small T antigen of BKV; JCV and MCV; and core, NS3 and NS5A of HCV have all been shown to drive oncogenic events through dysregulation of the activities of many cellular tumor suppressors. A partial list detailing the tumor viruses and their characteristics and association with different determinants driving these cancers is shown in Table 1. Included among these agents are HTLV-1 and HCV, which are RNA viruses. This is worth noting as virus infection can serve as oncogenic drivers but may not be independent or the sole drivers. Other contributory factors are also critical for progression of human cancers and include modulation of normal host regulatory events by interaction with the related host factors, mutations in key regulatory genes such as p53 and Rb, genetic susceptibility and exposure to hazardous carcinogens and pollutants.21
CELLULAR EVENTS TARGETED BY VIRAL-ENCODED ONCOGENES
Co-evolution of oncogenic viruses with their host enhances survival fitness and has equipped these viruses with ingenious ways to evade the host immune challenges as well as other regulatory molecular networks. Viruses can also disrupt cellular homeostasis by driving cell division, proliferation and apoptosis. Oncogenic viruses also encode homologs of cellular genes and have developed strategies of molecular mimicry such as expressing vBCL2 and vFLIP to subvert host cell division, survival, apoptosis and immune surveillance mechanisms.22 KSHV also encodes a number of genes, including vIL-6 (viral interleukin-6), vMIP-1 (viral macrophage inflammatory protein-1), vFLIP (viral FLICE inhibitory protein), vBCL-2, v-cyclin, vGPCR (viral G protein-coupled receptor) and vIRF-1 (viral interferon regulatory factor), which encodes similar functions to human cytokines and related signaling pathways.23 Interestingly, a monoclonal antibody raised in rabbit against the glycine-alanine domain of EBNA1 of EBV shows cross reactivity to a cellular host protein by recognizing the 72 kDa EBNA1 of EBV and a cellular protein of 62 kDa.24 Another example of molecular mimicry is LMP1 encoded by EBV, which mimics CD40 activities and recruits JAK3, TRADD and TRAF and bypass the NFκB signaling, evades apoptotic response and causes B-cell proliferation.25 Similarly, LMP2A targets host tyrosine kinase SYK and disrupts B-cell receptor (BCR) signaling.26 Typically, B cells without BCR activation undergo apoptotic death but LMP2A-expressing cells shuts down this apoptotic program through mimicking short linear motifs of the host protein.27
Current advances in cancer research have shown that, in parallel to genetic abnormalities, epigenetic modulation is a key factor for cancer development, causing activation of oncogenes, shutting off tumor-suppressor genes, suppressing DNA repair system and chromosomal instability, which leads to improper mitotic segregation.28
EBV latency programs restrict expression of EBV latency-associated genes in different types of cancers and are classified as latency 0–III.29 Removal of epigenetic marks either by histone deacetylase or a methylation inhibitor can switch between latency-associated gene expression patterns.30, 31 This implies that EBV latency programs are epigenetically regulated. EBV-encoded latency-associated gene products have a direct impact on the expression of host genes. EBNA2, an EBV nuclear antigen, interacts with p300, CBP and PCAF HATs through its acidic domain at the C-terminal. This suggests that EBNA2 can recruit these HATS to modulate host transcription.32 Furthermore, EBNA2 interaction with RBP-JK and PU.1 through the same domain in a sequence-specific manner is responsible for transcription activation of a number of host genes.32, 33
EBV-mediated global epigenetic alteration has been also shown in viral-positive nasopharyngeal carcinoma. These viral-encoded gene products are involved in methylation of tumor-suppressor genes in NPC.34 Specifically, the JNK-activator protein 1 signaling was shown to be exploited by EBV LMP1-mediated DNMT1 activation in NPC.34
THE FUNCTIONAL CONSEQUENCES OF NM23-H1 INTERACTION WITH CELLULAR AND VIRAL PARTNERS
Nm23-H1 is involved in regulation of many cellular functions. A model of its many representative functions is shown in Figure 2.
Schematic representation of four common functions of Nm23-H1. A primary function of Nm23-H1 is to maintain NTP homeostasis and supply GTP for signaling reactions. Other functions include DNase activity, histidine kinase activity and antimetastatic activity by regulating cellular functions, apoptosis, DNA repair, cell-cycle control and transcription regulation.
Cellular Interactions of Nm23-H1
Nm23 proteins interact with a large number of cellular proteins. We present a brief account of important interactions. Among these interactions, Nm23-H1, H-prune and cAMP phosphoesterase are well characterized. In several cancer types, H-prune shows augmented expression in colorectal, gastric cancer and breast cancer and has antimetastatic-suppressor activity.35 Interaction of H-prune with Nm23-H1 is facilitated by casein kinase 1 through serine residues at 120, 122 and 125 positions of Nm23-H1 and associated with enhanced neuroblastoma tumorigenesis.36, 37 H-prune association with Nm23-H1 is also reported as associated with the aggressive progression of gastric cancer and is associated with advanced grade of tumors.38
In association with cell motility and anchorage, gelsolin, an actin-binding protein, interacts with Nm23-H1 and Nm23-H1 dampens the actin-removing potential of gelsolin.39 Further, overexpression of gelsolin is associated with tumor metastasis by modulating cellular signaling cascades involving EGF and Ras-Rac.40, 41 Nm23-H1 also interacts with the serine–threonine kinase receptor associated protein (STRAP). This interaction downregulates transforming growth factor β (TGFβ) signaling.42, 43 Nm23-H1 interaction with STRAP stabilizes the binding of TGFβ and Smad7 and was also reported to activate the p53 tumor suppressor and its downstream effector functions, which includes cell-cycle regulation and induction of apoptosis.42, 43
Another important interaction of N23-H1 is with the kinase suppressor of Ras (KSR), a member of the MAP kinase pathway that contributes to metastasis.44 Further, the Rac1–GTPase interaction is regulated by Nm23-H1 and Tiam1 (T-cell lymphoma invasion and metastasis).45 Tiam1 is a known guanidine nucleotide exchange factor (GEF).
A member of the same family, the Dbl1 oncoprotein interacts with Nm23-H1 and leads to loss of the metastatic function of Nm23-H1.46 This happens through dampening the functional activity of Dbl1 as a GEF for Cdc42.46 Nm23-H1 also participates in GTPase regulation by interacting with Rad, a Ras-related protein associated with diabetes.47 Furthermore, direct interaction of Nm23-H1 with macrophage migratory and inhibitory factor (MIF) leads to loss of regulatory role of MIF in p53-mediated cell cycle regulation and apoptosis.48
In a recent study, the transcriptional factor forkhead box O3 (FOXO3) indirectly downregulates Nm23-H1 expression and enhances the metastatic event in non‐small‐cell lung cancer.49 Targeting FOXO3 as an upstream regulatory factor of Nm23-H1 may regulate its downstream antimetastatic function and provides an exciting opportunity for future therapeutic approaches.
The Enzymatic Activities of Nm23-H1
Nm23-H1 is the best-characterized member of Nm23 family and has three types of enzymatic activities associated.
Nucleoside diphosphatase kinase activity
NDPK activity of Nm23-H1 supplies the GTP for cellular signaling by transfer of phosphate between different nucleoside diphosphates and triphosphates.8, 50 Isoforms of Nm23 are differentially expressed in the tissue participate in the kinase activity.51 The primary function of its NDPK activity is to maintain NTP homeostasis for nucleic acid synthesis and to supply the energy for generating and maintaining cytosolic structure.52
Histidine kinase activity
Histidine protein kinases are well recognized in the microbial world as part of the two-component system and are absent in humans.53 Histidine kinases sense specific stimuli and the cognate response regulator manages the cellular response to that stimuli.53 Proteins phosphorylated at histidine residue by Nm23-H1 are ATP citrate lyase and succinate thiokinase and are linked with cell motility supression.54 Interestingly, serine/threonine phosphorylation of KSR by Nm23-H1 represents the direct association of the Nm23-H1 kinase activity and its role in regulating metastasis.44
DNase activity of Nm23-H1
Nm23-H1 remains as the part of SET complex activated by granzyme A.55 During apoptosis, Nm23-H1 localizes to the nucleus after degradation of the SET protein by granzyme to perform its nuclease activity. Granzyme A does not target Nm23-H1 rather it targets SET, and cytotoxic T-lymphocyte (CTL) attack can activate this process.55 This suggests that the tumorigenic suppression of Nm23-H1 will hinder the CTL-mediated immune attack and apoptotic events, allowing for tumor cells to bypass the cellular regulatory mechanisms. In in vitro assays, Nm23-H1 displayed the 3′–5′ exonuclease activity to single-stranded DNA substrates and double-stranded substrates with 3′ overhang but not with blunt-ended double-stranded substrates.56 Nm23-H1 nuclease activity links its metastatic-suppressor function with its exonuclease activity.57 Furthermore, Nm23-H1 overexpression in cancer cells suppresses the DNA repair activity, suggesting that Nm23-H1 is important for regulating the level of genomic instability in cancer.57
Viral Interactions with Nm23-H1 and its Related Functions
Nm23-H1 directly interacts with EBNA3C, the viral antigen encoded by EBV, through its carboxy-terminal domain as demonstrated via yeast two-hybrid screen where 365–992 amino acids were used as a bait to identify Nm23-H1.58 Findings were further validated where full length and the carboxy-terminal domain of EBNA3C interacted with Nm23-H1 with similar binding activities.58 In the presence of EBNA3C, Nm23-H1 can co-localize predominantly to the nuclear compartment with EBNA3C, translocating from the cytoplasm.58 In another study, breast carcinoma cells stably co-expressing EBNA3C and Nm23-H1 showed a higher rate of metastasis in nude mouse compared with EBNA3C and Nm23-H1 alone. This implies that EBV can overcome the antimetastatic effects of Nm23-H1 through its association with EBNA3C.59
Another essential EBV antigen EBNA1 is associated in molecular complex with Nm23-H1 as shown by co-immunoprecipitation using lymphoblastoid cells.60 The functional significance of Nm23-H1 association with EBNA1 was emphasized in the study of nude mice where EBNA1 promoted the in vivo growth of tumor cells.59 The antimetastatic effect of EBNA3 was, however, more potent in comparison to EBNA1.59
HPV, associated with anogenital, mouth and throat cancers, encodes two oncoproteins E6 and E7. These oncoproteins cause defect in cell division leading to chromosomal instability.61 The E7 is essential for HPV-mediated transformation of infected cells while E6 supports the process.62, 63 The E7 oncoprotein of HPV interacts with Nm23-H1 in yeast two-hybrid screen and was shown in cellular complex by co-immunoprecipitation.64 This interaction results in modulation of the transcription and translational activities of Nm23-H1 with direct functional consequences.64 Moreover, KSHV regulates Nm23-H1 through expression and interaction of the LANA, which activates the MAP kinase pathway and shifts the cellular location of Nm23-H1 to a predominantly nuclear compartment.65
HCV, an oncogenic RNA virus associated with liver cancer, encodes the E1 protein, which exhibits prometastatic activity. This activity is linked to the simultaneous downregulation of Nm23-H1 transcripts and its degradation. This results in the suppression of the functional activity of Nm23-H1.66
These examples give an overview of concerted molecular processes during viral infection where viral-encoded antigens drive the metastatic behavior of cells. EBNA3C encoded by EBV has potent prometastatic activity, and with its interaction with Nm23-H1, it regulates various downstream molecular events. Other viral antigens encoded by different oncoviruses such as LANA of KSHV and E7 of HPV drive similar potent prometastatic behavior. Thus this demonstrates that a complex cellular interaction web of host proteins are strategically tinkered by viral-encoded essential antigens that changes the specific promoter activity to modulate their expression, thereby changing the cellular behavior, as in the case of Necdin, where antiangiogenic activity is neutralized by the concerted action of EBNA3C and Nm23-H1.67
TRANSCRIPTION REGULATION OF CELLULAR TARGETS BY NM23 H1
Nm23-H1 is a significant cellular regulator that affects many signaling pathways related to tumorigenesis. For example, p53 can directly interact with Nm23-H1 at residues 113–290 in the DNA-binding region of p53.43 This interaction was mapped by mutational analysis and Nm23-H1 interaction with p53 along with its binding partner STRAP can positively influence p53 function related to arrest of cell cycle and induction of apoptosis.43 In reference to its contribution to angiogenic and invasive activities in breast cancer, the expression of Nm23-H1 and vascular endothelial growth factor (VEGF) can significantly modulate these functions.68
As a metastatic suppressor, Nm23-H1 regulates the expression of estrogen-responsive genes, including Bcl-2 through the direct interaction with estrogen receptor alpha, which has implications for its tumor-progression activities.69 Another study using immunohistochemistry followed by multivariate analysis for statistical association showed that PTEN, KAI-1 and Nm23-H1 downregulation strongly correlated with progression of non-small cell lung carcinomas.70 Further, KAI-1 expression was regulated through Nm23-H1 through modulation of KAI-1 promoter activity in L9981 lung cancer cells.71 Another study showed that c-Myc can upregulate the expression of Nm23-H1.72 Furthermore, the oncoprotein ErbB2 expression was shown to be regulated by Nm23-H1, which affects the cell proliferative activity of ErbB2.73
The transcriptional activity of Nm23-H1 was demonstrated using an Nm23-H1 fused to the Gal4 DNA-binding domain, which activates the upstream response element of a luciferase reporter construct.74 Co-transfection of EBNA3C further increased the luciferase activity.74 This established a transcriptional activation link between EBNA3C and Nm23-H1.74 Interestingly, cyclooxygenase-2 (Cox-2), an enzymatic mediator during conversion of arachidonate to prostaglandin and prostacyclin, that can promote inflammation is also regulated.75 Nm23-H1 modulates the promoter activity of Cox-2 and co-expression with EBNA3C further enhances the transcription effect of Nm23-H1.76 Thus Nm23-H1 can cooperatively contribute to the cellular inflammatory activity.76 Additionally, integrins are cell adhesion molecules shown to be associated with tumor development, progression and metastasis.77 Interestingly, Nm23-H1 and EBNA3C have contrasting effects on alpha v integrin expression as EBNA3C can upregulate alpha v integrin expression, and Nm23-H1 can downregulate its activity through targeted association with the Sp1 and GATA1 transcription factors.78
Tumor survival, invasion and metastatic activities are linked to the expression of metalloproteinases.79 Nm23-H1 cooperative interaction with EBNA3C changes the expression profile of metalloproteinase 9 (MMP9) and diminishes the antimetastatic effect of Nm23-H1 through targeting of the Ap-1 and NFκB transcription factors.80
Alteration in the glycosylation profiles can interfere with cellular immune surveillance with an impact on cancer.81, 82 Early expression of glycosyl transferases in cancer can serve as potential markers as seen in breast cancer.83 N-acetylglucosaminyltransferase V (GnT-V), a glycosyl transferase, shown to be regulated by Nm23-H1, can also have an impact on the metastatic phenotypes in lung cancer cells.84 Platelet-derived growth factor (PDGF) also contributed to angiogenesis, an important feature in cancer progression.85 Nm23-H1 suppressed the basal transcription of PDGF through modulation of its promoter regulatory elements through its DNA-binding activity.86
Further, Nm23-H1 was reported to be able to suppress the lysophosphatidic acid receptor EDG2 along with its antimetastatic action.87 Further, in studies involved with lung cancer cells, stable transfection of full-length Nm23-H1 in the non-small cell lung cancer cell line L9981 significantly heightened the expression of beta-Catenin, E-Cadherin and TIMP-1.88 This diminished the expression of MMP-2, CD44V6 and VEGF.88 The tissue inhibitor of metalloproteinases (Timp1) was shown to have antiproliferatory activity but, on overexpression in cancerous cells, mediated by aberrant glycosylation, led to loss of interaction with MMP and reversal of its related functional activity.88
These examples summarize a complex set of events where Nm23-H1 has direct influence on many cancer-related pathways, as a downstream effect of Nm23-H1 being an antimetastatic regulator. We can further conclude that if any upstream regulator such as viral factors exploits the Nm23-H1 they can directly or indirectly disrupt Nm23-H1 downstream effects, thereby shifting the behavior of the cell to be prometastatic.
NM23-H1 INTERACTION WITH EPIGENETIC MODIFIERS
Epigenetic silencing is a modality employed by many DNA viruses as well as other microbes for global modulation of host genes. This shifting of the epigenetic landscape can quickly suppress expression of tumor suppressors, activate oncogenes and alter the activities of epigenetic modifiers. One such example is viral-regulated epigenetic alteration associated with KSHV nuclear antigen LANA.89 Host DNA methyl transferases (DNMTs) were shown to be targeted by LANA.89 In vitro GST pull-down showed that recombinant LANA directly interacted with DNMT1, 3A and 3B.89 Furthermore, chromatin immunoprecipitation showed that the LANA–DNMT3A interaction was a prominent interaction in cells.89 Moreover, studies showed a direct correlation between LANA upregulation and nuclear Nm23-H1 leading to an increase in cellular invasion activity.65 Also, KSHV-encoded vIL-6 was shown to exploit the STAT3 pathway for genomic methylation by recruiting DNMT1 in endothelial cells.90
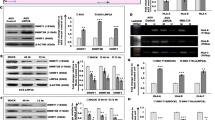
High-throughput ChIP sequencing has been employed to interrogate the EBV epigenome.91 A browser-based tool (https://ebv.wistar.upenn.edu/) is provided for open access to the raw alignment data and coverage track.91 These studies provide evidence for the global regulation of the viral genome. Recent studies in our laboratory showed that EBNA3C induced DNMT1 levels after EBV infection and can also interact with DNMT1 as shown by immunoprecipitation assays in EBNA3C-expressing cell lines (Figure 3a). Can Nm23-H1 and EBNA3C expression modulate the expression profile of different DNMTs as well as HDACs? To investigate this question, we now show that EBNA3C has a direct role in the regulation of DNMT1 levels as RT-PCR showed that the levels of DNMT 1 are more than doubled in Burkitt’s lymphoma cells stably expressing EBNA3C (Figure 3b, left panel). Furthermore, we also showed that the protein levels of DNMT1 were also dramatically increased in two independent cell lines expressing EBNA3C and that a lymphoblastoid cell line transformed by the virus EBV knocked down for EBNA3C showed a significant drop in levels of DNMT1.
EBV latent nuclear antigen 3C has a role in the regulation of DNMT1. (a) Immunoprecipitation of EBNA3C shows the presence of DNMT1. EBV-negative BJAB cells and BJAB7 and BJAB10 stably expressing EBNA3C were used for immunoprecipitation.104 Specific antibody for EBNA3C (A10) was used as previously described104 and anti-DNMT1 antibody was purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). (b) The left panel shows the RT-PCR for DNMT1 transcripts, and middle panel shows a western blotting analysis also showing increased DNMT1 protein levels. The right panel shows a WB with reduced DNMT1 levels when EBNA3C was knockdown by shRNA versus control shRNA. Expression of DNMT1 is higher in EBNA3C expressing BJAB-10 cells at the transcript level than control. At the protein level, there is a twofold higher expression of DNMT1 in EBNA3C-positive BJAB-7 and BJAB-10. Stable knockdown of EBNA3C in LCL1 cells significantly reduces the expression of DNMT1.
To determine the effect of NM23-H1 and EBNA3C on other DNMTs, we performed RT-PCR analysis on total RNAs from cells expressing Nm23-H1 alone or with EBNA3C. We showed that DNMT3a was strikingly upregulated with Nm23-H1 alone but was restored to the levels of vector control when EBNA3C was included (Figure 4a).
Effect of Nm23-H1 on Chromatin modifiers. (a) Effect of Nm23-H1 on the DNMT3a transcript is neutralized with simultaneous expression of EBNA3C. DNMT1 levels are significantly enhanced in the presence of Nm23-H1 and EBNA3C compared with Nm23-H1 alone. (b) HDAC1, 2 and 3 are enhanced with overexpression of Nm23-H1 but simultaneous expression of EBNA3C led to a reduction to normal transcription levels. (c) Nm23-H1 and EBNA3C co-expression enhanced DNMT1 expression compared with Nm23-H1 alone as seen by western blotting. Experiment was performed in MDA435 breast cancer cell line. Anti-Nm23-H1 antibody was purchased from Seikagaku Corp (Tokyo, Japan).105
The levels of HDAC 1, 2 and 3 were also increased on expression of Nm23-H1 but reduced to control levels when EBNA3C was expressed (Figure 4b). A drop in the levels of HDAC8 is seen with Nm23-H1, which was increased with EBNA3C (Figure 4b), suggesting that Nm23-H1 may be functioning in the downregulation of HDAC8 due to its role in suppressing apoptosis or other proliferative effects. Further studies using MDA-435 cells showed that, at the protein level, Nm23-H1 collaborating with EBNA3C can induce DNMT1 levels (Figure 4c).
PRMT5, a type II protein arginine methyl transferase, was also shown to have a direct role in regulating the cellular tumorigenic growth and proliferation event.92 It can negatively regulate the action of tumor suppressors as well as antiproliferative effects of Suppression of tumorigenicity 7 (ST7), Nm23-H1 and p107.92 ST7 and Nm23-H1 can form a complex with BRG1 and hBRM hSWI/SNF complexes, implying that this chromatin remodeling complex which includes PRMT5 may be involved in their regulation.92
Although this is a recent finding that EBV collaborates with Nm23-H1 for chromatin modification, it is on par with related functions of oncoviruses, including KSHV. This can be easily linked based on examples of other studies showing that modulating the host transcriptome and proteome has crucial role in the pathogenesis of virus-mediated cancers.
The detailed mechanism and signaling cascade is still not fully elucidated. However, epigenetic modification and microbial infection is not new. Chronic Helicobacter pylori infection in gastric cancer and nasopharyngeal carcinoma with EBV infection, which changes the large-scale epigenetic alteration and EBV linkage, are examples. Alterations in the host gene expression profiles can directly suppress host defenses against cancer.93, 94
POTENTIAL FOR THERAPEUTIC TARGETING
The expression and cytosolic accumulation compared with the nuclear localization of Nm23-H1 showed a reduction in Ras-MAP kinase activity resulting in loss of invasiveness in KSHV-mediated cancers.65 Similarly, removal of methylation marks from metastatic-compatible breast cancer cell lines also results in loss of proliferation and metastatic colonization activity by upregulation of Nm23-H1.95 This emphasized the therapeutic potential of Nm23-H1. Further, estrogen and progestin can also manipulate Nm23-H1 expression level to influence cell proliferation by activation of the PI3 kinase/Akt pathway as studied in ES-2, the ovarian cancer cell line.96 With regards to estrogen-induced accumulation of Nm23-H1, estrogen receptor alpha (ERα) is also increased and cells lacking the ERα have no effect on Nm23-H1 levels.97 It is suggested that the steroid hormones and their derivatives can positively regulate Nm23 expression, which results in a reduction in cell progression and metastasis.98, 99 Corticosteroids can also directly lead to an increase in Nm23-H1 levels through the glucocorticoid receptor.98, 99 A synthetic progesterone derivative, medoxyprogesterone acetate (MPA) in a mouse study model was shown to have the ability to increase the expression of Nm23-H1, reducing metastatic progression, and was suggested as a potential drug candidate for therapeutic targeting of Nm23-H1.98, 99 As a hormone therapy, MPA is at different stages of clinical trials for postmenopausal patients, including hormone receptor-positive breast cancer, and breast cancer of lobular, endometrium and duct as well as also in Paget’s disease of the bone.100
In prostate cancer, use of the competitive permeable peptide, which targets the Nm23-H1/hPrune interaction, leads to suppression of AKT/mTOR and NFκB pathways.101 In mouse xenograft models, a loss of metastatic activity was observed.101 Therefore, this has potential for therapeutic intervention as hPrune induction was observed in several cancer types, such as breast, esophageal squamous cell and gastric cancers.38, 102, 103 Further, a downstream target of Nm23-H1, lysophosphatidic acid receptor 1 (LPA1) if inhibited can have therapeutic implications as the expression of LPA1 was suppressed by Nm23-H1 induction with antimetastatic effects.87
References
Fidler IJ . The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nat Rev Cancer 2003;3:453–458.
Paget S . The distribution of secondary growth in cancer of breast. Lancet 1889;133:571–573.
Gupta GP, Massagué J . Cancer metastasis: building a framework. Cell 2006;127:679–695.
Horak CE, Lee JH, Marshall JC et al. The role of metastasis suppressor genes in metastatic dormancy. APMIS 2008;116:586–601.
Steeg PS, Bevilacqua G, Kopper L et al. Evidence for a novel gene associated with low tumor metastatic potential. J Natl Cancer Inst 1988;80:200–204.
Stahl JA, Leone A, Rosengard AM et al. Identification of a second human nm23 gene, nm23-H2. Cancer Res 1991;51:445–449.
Tong Y, Yung LY, Wong YH . Metastasis suppressors Nm23H1 and Nm23H2 differentially regulate neoplastic transformation and tumorigenesis. Cancer Lett 2015;361:207–217.
Berg JM, Tymoczko JL, Stryer L . Biochemistry. WH Freeman: New York, USA, 2000.
The Nobel Prize in Physiology or Medicine 1989, [Internet]. The Noble Assembly at the Karolinska Institute, Stockholm (Sweden), 1989. Available from http://www.nobelprize.org/nobel_prizes/medicine/laureates/1989/press.hyml.
Shu X-S, Li L, Tao Q . Chromatin regulators with tumor suppressor properties and their alterations in human cancers. Epigenomics 2012;4:537–549.
Zhang P, Torres K, Liu X et al. An overview of chromatin-regulating proteins in cells. Curr Protein Pept Sci 2016;17:401–410.
Clapier CR, Cairns BR . The biology of chromatin remodeling complexes. Ann Rev Biochem 2009;78:273–304.
Narlikar GJ, Sundaramoorthy R, Owen-Hughes T . Mechanisms and functions of ATP-dependent chromatin-remodeling enzymes. Cell 2013;154:490–503.
Bannister AJ, Kouzarides T . Regulation of chromatin by histone modifications. Cell Res 2011;21:381–395.
Choudhary C, Weinert BT, Nishida Y et al. The growing landscape of lysine acetylation links metabolism and cell signalling. Nat Rev Mol Cell Biol 2014;15:536–550.
Zentner GE, Henikoff S . Regulation of nucleosome dynamics by histone modifications. Nat Struct Mol Biol 2013;20:259–266.
McGrath J, Trojer P . Targeting histone lysine methylation in cancer. Pharmacol Ther 2015;150:1–22.
Bedford MT, Clarke SG . Protein arginine methylation in mammals: who, what, and why. Mol Cell 2009;33:1–13.
Anand P, Kunnumakara AB, Sundaram C et al. Cancer is a preventable disease that requires major lifestyle changes. Pharm Res 2008;25:2097–2116.
Vandeven N, Nghiem P . Pathogen-driven cancers and emerging immune therapeutic strategies. Cancer Immunol Res 2014;2:9–14.
Liao JB . Viruses and human cancer. Yale J Biol Med 2006;79:115–122.
Kvansakul M, Hinds MG . Structural biology of the Bcl-2 family and its mimicry by viral proteins. Cell Death Dis 2013;4:e909.
Moore PS, Boshoff C, Weiss RA et al. Molecular mimicry of human cytokine and cytokine response pathway genes by KSHV. Science 1996;274:1739–1744.
Luka J, Kreofsky T, Pearson GR et al. Identification and characterization of a cellular protein that cross-reacts with the Epstein-Barr virus nuclear antigen. J Virol 1984;52:833–838.
Thorley-Lawson DA . Epstein-Barr virus: exploiting the immune system. Nat Rev Immunol 2001;1:75–82.
Fruehling S, Longnecker R . The immunoreceptor tyrosine-based activation motif of Epstein-Barr virus LMP2A is essential for blocking BCR-mediated signal transduction. Virology 1997;235:241–251.
Merchant M, Caldwell RG, Longnecker R . The LMP2A ITAM is essential for providing B cells with development and survival signals in vivo. J Virol 2000;74:9115–9124.
Jones PA, Baylin SB . The fundamental role of epigenetic events in cancer. Nat Rev Genet 2002;3:415–428.
Young LS, Murray PG . Epstein–Barr virus and oncogenesis: from latent genes to tumours. Oncogene 2003;22:5108–5121.
Masucci MG, Contreras-Salazar B, Ragnar E et al. 5-Azacytidine up regulates the expression of Epstein-Barr virus nuclear antigen 2 (EBNA-2) through EBNA-6 and latent membrane protein in the Burkitt’s lymphoma line rael. J Virol 1989;63:3135–3141.
Nishikawa J, Kis LL, Liu A et al. Upregulation of LMP1 expression by histone deacetylase inhibitors in an EBV carrying NPC cell line. Virus Genes 2004;28:121–128.
Wang L, Grossman SR, Kieff E . Epstein-Barr virus nuclear protein 2 interacts with p300, CBP, and PCAF histone acetyltransferases in activation of the LMP1 promoter. Proc Natl Acad Sci USA 2000;97:430–435.
Grossman SR, Johannsen E, Tong X et al. The Epstein-Barr virus nuclear antigen 2 transactivator is directed to response elements by the J kappa recombination signal binding protein. Proc Natl Acad Sci USA 1994;91:7568–7572.
Tsai CL, Li HP, Lu YJ et al. Activation of DNA methyltransferase 1 by EBV LMP1 involves c-Jun NH 2-terminal kinase signaling. Cancer Res 2006;66:11668–11676.
Forus A, D’Angelo A, Henriksen J et al. Amplification and overexpression of PRUNE in human sarcomas and breast carcinomas-a possible mechanism for altering the nm23-H1 activity. Oncogene 2001;20:6881–6890.
Garzia L, Roma C, Tata N et al. H-prune-nm23-H1 protein complex and correlation to pathways in cancer metastasis. J Bioenerg Biomembr 2006;38:205–213.
Carotenuto M, Pedone E, Diana D et al. Neuroblastoma tumorigenesis is regulated through the Nm23-H1/h-Prune C-terminal interaction. Sci Rep 2013;3:1351.
Oue N, Yoshida K, Noguchi T et al. Increased expression of h-prune is associated with tumor progression and poor survival in gastric cancer. Cancer Sci 2007;98:1198–1205.
Marino N, Marshall JC, Collins JW et al. Nm23-H1 binds to Gelsolin and inactivates its actin-severing capacity to promote tumor cell motility and metastasis. Cancer Res 2013;73:5949–5962.
Lader AS, Lee JJ, Cicchetti G et al. Mechanisms of gelsolin-dependent and -independent EGF-stimulated cell motility in a human lung epithelial cell line. Exp Cell Res 2005;307:153–163.
De Corte V, Bruyneel E, Boucherie C et al. Gelsolin-induced epithelial cell invasion is dependent on Ras-Rac signaling. EMBO J 2002;21:6781–6790.
Seong H-A, Jung H, Choi H-S et al. Regulation of transforming growth factor-beta signaling and PDK1 kinase activity by physical interaction between PDK1 and serine-threonine kinase receptor-associated protein. J Biol Chem 2005;280:42897–42908.
Jung H, Seong HA, Ha H . NM23-H1 tumor suppressor and its interacting partner STRAP activate p53 function. J Biol Chem 2007;282:35293–35307.
Hartsough MT, Morrison DK, Salerno M et al. Nm23-H1 metastasis suppressor phosphorylation of kinase suppressor of Ras via a histidine protein kinase pathway. J Biol Chem 2002;277:32389–32399.
Otsuki Y, Tanaka M, Yoshii S et al. Tumor metastasis suppressor nm23H1 regulates Rac1 GTPase by interaction with Tiam1. Proc Natl Acad Sci USA 2001;98:4385–4390.
Murakami M, Meneses PI, Knight JS et al. Nm23-H1 modulates the activity of the guanine exchange factor Dbl-1. Int J Cancer 2008;123:500–510.
Zhu J, Tseng YH, Kantor JD et al. Interaction of the Ras-related protein associated with diabetes rad and the putative tumor metastasis suppressor NM23 provides a novel mechanism of GTPase regulation. Proc Natl Acad Sci USA 1999;96:14911–14918.
Jung H, Seong HA, Ha H . Direct interaction between NM23-H1 and macrophage migration inhibitory factor (MIF) is critical for alleviation of MIF-mediated suppression of p53 activity. J Biol Chem 2008;283:32669–32679.
Zhang L, Li L, Wei H et al. Transcriptional factor FOXO3 negatively regulates the expression of nm23-H1 in non-small cell lung cancer. Thorac Cancer 2016;7:9–16.
Kim HD, Youn B, Kim TS et al. Regulators affecting the metastasis suppressor activity of Nm23-H1. Mol Cell Biochem 2009;329:167–173.
Lacombe ML, Milon L, Munier A et al. The human Nm23/nucleoside diphosphate kinases. J Bioenerg Biomembr 2000;32:247–258.
Kaul R, Murakami M, Kumar P et al, Nm23 as a metastatic inhibitor In: Thomas-Tikhonenko A (ed). Cancer Genome and Tumor Microenvironment. Springer: New York, NY, USA, 2010, pp 233–271.
Stock AM, Robinson VL, Goudreau PN . Two-component signal transduction. Annu Rev Biochem 2000;69:183–215.
Wagner PD, Steeg PS, Vu ND . Two-component kinase-like activity of nm23 correlates with its motility-suppressing activity. Proc Natl Acad Sci USA 1997;94:9000–9005.
Fan Z, Beresford PJ, Oh DY et al. Tumor suppressor NM23-H1 is a granzyme A-activated DNase during CTL-mediated apoptosis, and the nucleosome assembly protein set is its inhibitor. Cell 2003;112:659–672.
Ma D, McCorkle JR, Kaetzel DM . The metastasis suppressor NM23-H1 possesses 3’-5’ exonuclease activity. J Biol Chem 2004;279:18073–18084.
Kaetzel DM, Zhang Q, Yang M et al. Potential roles of 3′-5′exonuclease activity of NM23-H1 in DNA repair and malignant progression. J Bioenerg Biomembr 2006;38:163–167.
Subramanian C, Cotter Ma, Robertson ES . Epstein-Barr virus nuclear protein EBNA-3C interacts with the human metastatic suppressor Nm23-H1: a molecular link to cancer metastasis. Nat Med 2001;7:350–355.
Kaul R, Murakami M, Choudhuri T et al. Epstein-Barr virus latent nuclear antigens can induce metastasis in a nude mouse model. J Virol 2007;81:10352–10361.
Murakami M, Lan K, Subramanian C et al. Epstein-Barr virus nuclear antigen 1 interacts with Nm23-H1 in lymphoblastoid cell lines and inhibits its ability to suppress cell migration. J Virol 2005;79:1559–1568.
Duensing S, Lee LY, Duensing A et al. The human papillomavirus type 16 E6 and E7 oncoproteins cooperate to induce mitotic defects and genomic instability by uncoupling centrosome duplication from the cell division cycle. Proc Natl Acad Sci USA 2000;97:10002–10007.
Munger K, Basile JR, Duensing S et al. Biological activities and molecular targets of the human papillomavirus E7 oncoprotein. Oncogene 2001;20:7888–7898.
Zwerschke W, Jansen-Durr P . Cell transformation by the E7 oncoprotein of human papillomavirus type 16: interactions with nuclear and cytoplasmic target proteins. AdvCancer Res 2000;78:1–29.
Mileo AM, Piombino E, Severino A et al. Multiple interference of the human papillomavirus-16 E7 oncoprotein with the functional role of the metastasis suppressor Nm23-H1 protein. J Bioenerg Biomembr 2006;38:215–225.
Qin Z, Dai L, Toole B et al. Regulation of Nm23-H1 and cell invasiveness by Kaposi’s sarcoma-associated herpesvirus. J Virol 2011;85:3596–3606.
Khera L, Paul C, Kaul R . Hepatitis C Virus E1 protein promotes cell migration and invasion by modulating cellular metastasis suppressor Nm23-H1. Virology 2017;506:110–120.
Saha A, Robertson ES . Functional modulation of the metastatic suppressor Nm23-H1 by oncogenic viruses. FEBS Lett 2011;585:3174–3184.
Huang ZH, Su GQ, Mao QG et al. Expressions of vascular endothelial growth factor and nm23-H1 gene and their relation to the prognosis of breast cancer in young women. Di Yi Jun Yi Da Xue Xue Bao 2004;24:1398–1401.
Curtis CD, Likhite VS, McLeod IX et al. Interaction of the tumor metastasis suppressor nonmetastatic protein 23 homologue H1 and estrogen receptor alpha alters estrogen-responsive gene expression. Cancer Res 2007;67:10600–10607.
Goncharuk VN, Del-Rosario A, Kren L et al. Co-downregulation of PTEN, KAI-1, and nm23-H1 tumor/metastasis suppressor proteins in non-small cell lung cancer. Ann Diagn Pathol 2004;8:6–16.
You J, Chang R, Liu B et al. Nm23-H1 was involved in regulation of KAI1 expression in high-metastatic lung cancer cells L9981. J Thorac Dis 2016;8:1217–1226.
Godfried MB, Veenstra M, v Sluis P et al. The N-myc and c-myc downstream pathways include the chromosome 17q genes nm23-H1 and nm23-H2. Oncogene 2002;21:2097–2101.
Tommasi S, Fedele V, Crapolicchio A et al. ErbB2 and the antimetastatic nm23/NDP kinase in regulating serum induced breast cancer invasion. Int J Mol Med 2003;12:131–134.
Subramanian C, Robertson ES . The metastatic suppressor Nm23-H1 interacts with EBNA3C at sequences located between the glutamine- and proline-rich domains and can cooperate in activation of transcription. J Virol 2002;76:8702–8709.
Ricciotti E, Fitzgerald GA . Prostaglandins and inflammation. Arterioscler Thromb Vasc Biol 2011;31:986–1000.
Kaul R, Verma SC, Murakami M et al. Epstein-Barr virus protein can upregulate cyclo-oxygenase-2 expression through association with the suppressor of metastasis Nm23-H1. J Virol 2006;80:1321–1331.
Desgrosellier JS, Cheresh DA . Integrins in cancer: biological implications and therapeutic opportunities. Nat Rev Cancer 2010;10:9–22.
Choudhuri T, Verma SC, Lan K et al. Expression of alpha V integrin is modulated by Epstein-Barr virus nuclear antigen 3C and the metastasis suppressor Nm23-H1 through interaction with the GATA-1 and Sp1 transcription factors. Virology 2006;351:58–72.
Stamenkovic I . Matrix metalloproteinases in tumor invasion and metastasis. Semin Cancer Biol 2000;10:415–433.
Kuppers DA, Lan K, Knight JS et al. Regulation of matrix metalloproteinase 9 expression by Epstein-Barr virus nuclear antigen 3C and the suppressor of metastasis Nm23-H1. J Virol 2005;79:9714–9724.
Hakomori SI . Aberrant glycosylation in cancer cell membranes as focused on glycolipids: overview and perspectives. Cancer Res 1985;45:2405–2414.
Madsen CB, Petersen C, Lavrsen K et al. Cancer associated aberrant protein O-glycosylation can modify antigen processing and immune response. PLoS ONE 2012;7:e50139.
Andergassen U, Liesche F, Kölbl AC et al. Glycosyltransferases as markers for early tumorigenesis. Biomed Res Int 2015;2015:1–11.
Guo HB, Liu F, Zhao JH et al. Down-regulation of N-acetylglucosaminyltransferase V by tumorigenesis- or metastasis-suppressor gene and its relation to metastatic potential of human hepatocarcinoma cells. J Cell Biochem 2000;79:370–385.
Ariad S, Seymour L, Bezwoda WR . Platelet-derived growth factor (PDGF) in plasma of breast cancer patients: correlation with stage and rate of progression. Breast Cancer Res Treat 1991;20:11–17.
Ma D, Xing Z, Liu B et al. NM23-H1 and NM23-H2 repress transcriptional activities of nuclease-hypersensitive elements in the platelet-derived growth factor-A promoter. J Biol Chem 2002;277:1560–1567.
Horak CE, Mendoza A, Vega-Valle E et al. Nm23-H1 suppresses metastasis by inhibiting expression of the lysophosphatidic acid receptor EDG2. Cancer Res 2007; 6711751–6711759.
Kim YS, Kim SH, Kang JG et al. Expression level and glycan dynamics determine the net effects of timp-1 on cancer progression. BMB Rep 2012;45:623–628.
Shamay M, Krithivas A, Zhang J et al. Recruitment of the de novo DNA methyltransferase Dnmt3a by Kaposi’s sarcoma-associated herpesvirus LANA. Proc Natl Acad Sci USA 2006;103:14554–14559.
Wu J, Xu Y, Mo D et al. Kaposi’s Sarcoma-Associated Herpesvirus (KSHV) vIL-6 promotes cell proliferation and migration by upregulating DNMT1 via STAT3 activation. PLoS ONE 2014;9:e93478.
Arvey A, Tempera I, Lieberman PM . Interpreting the Epstein-Barr virus (EBV) epigenome using high-throughput data. Viruses 2013;5:1042–1054.
Pal S, Vishwanath SN, Erdjument-Bromage H et al. Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol Cell Biol 2004;24:9630–9645.
Hanash S, Schliekelman M . Proteomic profiling of the tumor microenvironment: recent insights and the search for biomarkers. Genome Med 2014;6:12.
Swann JB, Smyth MJ . Immune surveillance of tumors. J Clin Invest 2007;117:1137–1146.
Hartsough MT, Clare SE, Mair M et al. Elevation of breast carcinoma Nm23-H1 metastasis suppressor gene expression and reduced motility by DNA methylation inhibition. Cancer Res 2001;61:2320–2327.
Hua K-Q, Yao L-Q, Cao Q et al. Influence of estrogen and progestin on nm23-H1 expression in epithelial ovarian cancer cell lines via activation of phosphorylation signaling. Zhonghua Fu Chan Ke Za Zhi 2006;41:756–761.
Lin K-H, Wang W-J, Y-H WU et al. Activation of antimetastatic Nm23-H1 gene expression by estrogen and its alpha-receptor. Endocrinology 2002;143:467–475.
Palmiere D, Halverson DO, Ouatas T et al. Medroxyprogesterone acetate elevation of Nm23-H1 metastasis suppressor expression in hormone receptor-negative breast cancer. J Natl Cancer Inst 2005;97:632–642.
Palmieri D, Horak CE, Lee J-H et al. Translational approaches using metastasis suppressor genes. J Bioenerg Biomembr 2006;38:151–161.
Sherbet GV Metastasis suppressor nm23 and manipulation of its expression. In: Therapeutic Strategies in Cancer Biology and Pathology. Elsevier: Waltham, MA, USA, 2013, pp 83–87.
Carotenuto M, De Antonellis P, Chiarolla CM et al. A therapeutic approach to treat prostate cancer by targeting Nm23-H1/h-Prune interaction. Naunyn Schmiedebergs Arch. Pharmacol 2014;388:257–269.
Zollo M, Andrè A, Cossu A et al. Overexpression of h-prune in breast cancer is correlated with advanced disease status. Clin Cancer Res 2005;11:199–205.
Noguchi T, Oue N, Wada S et al. h-Prune is an independent prognostic marker for survival in esophageal squamous cell carcinoma. Ann Surg Oncol 2009;16:1390–1396.
Pei Y, Banerjee S, Sun Z et al. EBV nuclear antigen 3C mediates regulation of E2F6 to inhibit E2F1 transcription and promote cell proliferation. PLoS Pathog 2016;12:1–24.
Murakami M, Meneses PI, Lan K et al. The suppressor of metastasis Nm23-H1 interacts with the Cdc42 Rho family member and the pleckstrin homology domain of oncoprotein Dbl-1 to suppress cell migration. Cancer Biol Ther 2008;7:677–688.
Acknowledgements
The work was supported by R01-CA171971-01A1, R01-CA177423-01 and P01-CA174439-02 grants to ESR.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
In this review, Nm23-H1-mediated cellular and viral interactions with an emphasis on chromatin modifications are presented. How Nm23-H1 modulates the activities of chromatin modifiers through interaction with Epstein-Barr virus-encoded oncogenic antigens and related crosstalk is discussed. Potential therapeutic approaches targeting Nm23-H1 are also outlined.
Rights and permissions
About this article
Cite this article
Pandey, S., Robertson, E. Oncogenic Epstein–Barr virus recruits Nm23-H1 to regulate chromatin modifiers. Lab Invest 98, 258–268 (2018). https://doi.org/10.1038/labinvest.2017.112
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/labinvest.2017.112
This article is cited by
-
The roles of DNA methylation on the promotor of the Epstein–Barr virus (EBV) gene and the genome in patients with EBV-associated diseases
Applied Microbiology and Biotechnology (2022)
-
The NDPK/NME superfamily: state of the art
Laboratory Investigation (2018)