Abstract
The construction of tissue microarrays (TMAs) with cores from a large number of paraffin-embedded tissues (donors) into a single paraffin block (recipient) is an effective method of analyzing samples from many patient specimens simultaneously. For the TMA to be successful, the cores within it must capture the correct histologic areas from the donor blocks (technical accuracy) and maintain concordance with the tissue of origin (analytical accuracy). This can be particularly challenging for tissues with small histological features such as small islands of carcinoma in situ (CIS), thin layers of normal urothelial lining of the bladder, or cancers that exhibit intratumor heterogeneity. In an effort to create a comprehensive TMA of a bladder cancer patient cohort that accurately represents the tumor heterogeneity and captures the small features of normal and CIS, we determined how core size (0.6 vs 1.0 mm) impacted the technical and analytical accuracy of the TMA. The larger 1.0 mm core exhibited better technical accuracy for all tissue types at 80.9% (normal), 94.2% (tumor), and 71.4% (CIS) compared with 58.6%, 85.9%, and 63.8% for 0.6 mm cores. Although the 1.0 mm core provided better tissue capture, increasing the number of replicates from two to three allowed with the 0.6 mm core compensated for this reduced technical accuracy. However, quantitative image analysis of proliferation using both Ki67+ immunofluorescence counts and manual mitotic counts demonstrated that the 1.0 mm core size also exhibited significantly greater analytical accuracy (P=0.004 and 0.035, respectively, r2=0.979 and 0.669, respectively). Ultimately, our findings demonstrate that capturing two or more 1.0 mm cores for TMA construction provides superior technical and analytical accuracy over the smaller 0.6 mm cores, especially for tissues harboring small histological features or substantial heterogeneity.
Similar content being viewed by others
Main
Tissue microarrays (TMAs) are constructed by transferring selected cores from a large number of standard formalin-fixed paraffin-embedded tissues (donor blocks) into a single paraffin block (recipient block). The technique was first introduced by Dr Battifora who prepared blocks by wrapping tissue samples from different organs in a sheet of small intestine and embedded this cylinder of tissue in a single paraffin block.1 The TMA became an established technique after Kononen et al2 developed an array-based, high-throughput method that facilitated simultaneous analysis of a very large number of breast tumors.
Modern TMAs are constructed with cores ranging from 0.6 to 2.0 mm in diameter and each tissue is represented by one to four cores per patient. The larger core size increases the amount of tissue represented in the TMA but decreases the TMA density (#cores/TMA block, see Table 1). Consequently, the number of patients included in a single block corresponds directly to the size of the core and the quality of the TMA depends largely on the representation of patient tissue captured in each core. Nevertheless, good concordance of TMA cores with the original whole-tissue section can be achieved3 and this has made TMAs a highly desirable resource. Not only do they greatly reduce the labor, time, and reagents needed for a single study, parallel analysis in a TMA can also improve accuracy by reducing batch-to-batch variability and improved statistical assessment of a large patient cohort. Moreover, TMAs readily integrate advanced image analysis approaches that are developed for complex multi-parametric investigations.
With new automated technologies accelerating their production,4 TMAs are rapidly adopted in many environments. However, a number of challenges must be considered when attempting to use a TMA for tissue-based studies of a large patient cohort: (1) The histological feature of interest may be small and difficult to capture within a TMA punch whose placement itself is variable.5 (2) Any single core taken from heterogeneous tissue may be unable to accurately reflect the molecular complexity of the donor tissue. (3) Quantitative analysis of the staining may require multiple cores to enable appropriate statistical assessment. Fortunately, previous histological analyses of the proliferation marker Ki67 in a bladder TMA has shown that a TMA can accurately capture intratumor heterogeneity.6 Moreover, selecting cores from different areas of the block (center, periphery, invasive, or noninvasive) or, sometimes, different donor blocks can also reduce the scope of these problems and produce TMAs with good representation of the original tissue.7, 8
Some investigators have concluded that core size was not relevant,9 whereas others have preferred cores up to 2.0 mm in diameter, particularly when comparing different histological zones.5, 10, 11 However, a number of studies comparing distinct tissues such as tumor, normal, and lymph node metastasis, used 1.0 mm cores5 with the assumption that this larger core size increase the accuracy of punching the smaller regions of interest.11 To compensate for the limited accuracy of smaller core size, the number of cores representing a histological zone can be increased but there is no published consensus on what that number should be. Studies of breast ductal carcinoma in situ (DCIS) and pleomorphic adenoma have been successful with two cores,10, 12 whereas others have preferred three cores/tissue.13, 14, 15, 16 Adjusting both the number and the size of TMA cores can be beneficial as shown by Remotti11 who used a combination of up to four cores of 1.5 mm or 2.0 mm to achieve 95–97% concordance. Unfortunately, increasing the number of cores included in a TMA decreases the density of patients represented within such a block (Table 1) and increasing the core size accelerates depletion of the donor block. Both of these are contrary to the fundamental advantage of a TMA and a balance must be struck between the core size and the number of cores/tissue/patient to maximize the accuracy of the TMA and still gain the benefit of large-scale parallel analysis of many tissue specimens. Ultimately, TMAs with smaller core sizes and a smaller number of cores/tissue have greater patient density and reduce depletion of the donor tissue. Conversely, TMAs with larger cores and/or a greater number of cores can be more accurate. Consequently, an informed TMA-design requires careful consideration of both core size and the number of cores/tissue/patient to optimize the balance between scope of the TMA and its accuracy.
Constructing a TMA of bladder cancer has its own unique challenges. The tumor histology is variable and shows multiple morphologic features including fungating masses projecting out of the bladder mucosa (ie, papillary lesions), regionally differentiating subtypes (ie, squamous differentiation), local tissue responses (desmoplastic reactions and inflammation) and invasive tumors penetrating deep into the muscle and adipose tissues. Moreover, many bladder cases present as noninvasive masses17 or the tumor is visible only as a malignant change in the surface epithelium such as in CIS. Including normal bladder urothelium is also difficult because it comprises only a few cell layers making it extremely difficult to accurately capture this tissue in a small core. Although most bladder TMAs report reasonable concordance with the donor tumor tissue, there are no reports on the TMA accuracy for smaller features such as CIS and normal urothelium. As a 1.0 mm core has nearly three times the surface area of a 0.6 mm core (Figure 1), this larger core should theoretically be more effective at capturing the small histological features and thereby enhance the TMA’s technical accuracy. Moreover, we hypothesized that the larger cores would exhibit less inter-core variability and better concordance with the donor block which, in turn, should improve the TMA’s analytical accuracy.
Tissue microarray construction. (a) Slides containing the desired target tissue were digitally scanned and annotated using the 3D Histotech system. Cores were extracted from donor blocks using an automated arrayer guided by the annotated digital slides. Either nine (TMA1) or six (TMA2) cores were transferred to corresponding recipient blocks. (b) Representation of the core size, core area, and an H&E of TMA1 and TMA2.
To determine which core size was most appropriate for the production of a bladder cancer TMA, we performed a direct comparison of 0.6 mm and 1.0 mm core sizes. Ultimately, the 1.0 mm core size provided better technical and analytical accuracy. Increasing the number of 0.6 mm cores could compensate partially for the lower technical accuracy of this core size but not for the lower analytical accuracy. Our findings suggest that a TMA block constructed with two 1.0 mm cores for each tissue is optimal compared with three 0.6 mm cores. In addition, we recommend that each TMA block is produced in duplicate or triplicate such that more than two cores are available for advanced statistical analyses. (Can be used as a summary).
Materials and methods
TMA Construction
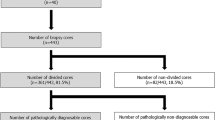
Two pilot TMAs were constructed with 0.6 and 1.0 mm diameter core size, respectively. In TMA1, each patient is represented by three 0.6 mm cores/tissue in three replicate blocks (three cores/block=9 cores total). In TMA2, each patient is represented by two 1.0 mm cores/tissue in three replicate blocks (two cores/block=6 cores total).
Cases were selected from the archive of the Vanderbilt University Medical Center, from years 2000–2012. Approval for the study was granted by the institution review board (IRB# 150278). Study data were collected and managed using REDCap18 electronic data capture tools hosted at Vanderbilt University Medical Center. Each case included at least two different bladder histologic tissue types (normal, tumor, and/or Cis). Diagnostic hematoxylin and eosin (H&E) slides from each patient were reviewed by a trained pathologist to identify the target tissue. The slides of selected donor blocks were digitized and placement of the cores in each histologic tissue type were annotated in Panoramic Viewer (3D Histech, Budapest, Hungary) by a trained pathologist. Annotated cores from the donor block were robotically transferred to the recipient TMA block using the 3D Histech automated tissue microarrayer (3D Histech).19 A map was generated specifying the exact position of each core and cores were linked to their donor and corresponding clinical data. This facilitated image export and assessment of TMA sections. Control cores were generated from the tonsils, liver, and kidney tissue, as well as from the cell pellets of bladder cancer cell lines. Individual TMA block sections were cut at 5 μm. The first and the fifth sections were stained with H&E for pathology assessment. Whole slide imaging and quantification of immunostaining were performed in the Digital Histology Shared Resource at Vanderbilt University Medical Center (www.mc.vanderbilt.edu/dhsr).
Technical Accuracy Assessment
A scoring system was designed to evaluate how accurately the target tissue was captured in each of the cores (Figure 2). Each core was given a score of 0 through 3 depending on whether the core contained ≥50% of target tissue (score=3), <50% of target tissue (score=2), no target tissue (score=1), or no tissue at all (score=0). One H&E stained section from each TMA paraffin block (one from each of the triplicates) was used for technical assessment. Cores with technical scores 2 or 3 were considered usable, whereas those with 0 or 1 were considered non-usable. For each tissue, the overall quality of captured material was defined as the mean score of the usable cores. A patient’s material was considered analyzable if at least two cores were considered ‘Usable’ by the above scoring. Chi-square and Fisher’s exact tests were used to determine the difference in proportions of usable to non-usable cases between TMA groups. See also Table 2.
Analytical Accuracy Assessment
The ability to analyze each core was evaluated using both immunofluorescence staining of Ki67 and manual mitotic spindle counts. One section from TMA1 (0.6 mm cores) and TMA2 (1.0 mm cores) was stained for collagen (CNA35),20 Ki67 (SP6-Thermo-Scientific, 1:500), and Hoechst (nuclear counterstain). The stained slides were scanned at × 20 magnification with an Ariol SL-50 (Leica Biosystems) and composite images for each individual core were batch exported as three-color, red/green/blue (RGB) jpeg2000 files. The images were then batch converted to Portable Network Graphics (png) format using the software Automator (Apple Inc.). Ki67+ nuclei were quantified in ImageJ21 (http://imagej.nih.gov/ij/) by splitting each image into the three individual color channels, manually thresholding the red (Ki67) and blue (nuclei) channels, segmenting the objects with ‘watershed’, and counting objects with the ‘analyze particles’ function. Analytical accuracy was assessed by determining the inter-core variability (replicate core standard deviation) and the concordance (Pearson correlation coefficient) between the mean Ki67+ nuclei in the replicate cores and the Ki67+ nuclei in their corresponding donor whole sections (n=6). Analytical accuracy was also determined by correlating the mean number of mitotic figures in 10 high-power fields, counted manually by a pathologist, in H&E whole-tissue sections with the mean Ki67+ nuclei in corresponding replicate TMA cores.
Results
TMA Construction
Using 0.6 and 1.0 mm cores, it was possible to construct TMAs with core densities of 400 (16 × 25) and 198 (11 × 18), respectively. The automated arrayer (Supplementary Figure 1) was used to select nine 0.6 mm cores or six 1.0 mm cores from each donor tissue and distribute them across three replicate TMA recipient blocks. This created each TMA in triplicate with three cores/tissue/block in TMA1 (0.6 mm) and two cores/tissue/block in TMA2 (1.0 mm). Based on these dimensions, each patient sample was represented by 0.283 mm2 (TMA1) or 0.785 mm2 (TMA2) in each TMA block (Figure 1).
Technical Accuracy
Hematoxylin and eosin (H&E) stained slides from each triplicate recipient block of TMA1 (0.6 mm) and TMA2 (1.0 mm) were assessed for their ability to capture the desired target tissue within the core using the scoring system described in the 'Materials and methods' section and in Figure 2. A comparison of usable cores for tumor, normal, and CIS tissue within each TMA (0.6 and 1.0 mm, Table 2) revealed that the technical accuracy was worse for the small histological features of normal urothelium and CIS tissue than the larger tumor tissues, in both TMA1 and TMA2 (tumor vs normal P≤0.0001, tumor vs CIS P≤0.0001). In addition, the larger 1.0 mm core size (TMA2) had a significantly greater percentage of usable cores for normal and tumor tissue in comparison with the smaller 0.6 mm core size (tumor, 94.2% vs 84.9%, P=0.004 and normal, 80.9% vs 58%, P=0.0007; Table 3). A significant difference could not be reached for the CIS cores owing to the limited number of CIS cases in both the TMAs. Although the number of usable cores (score ≥2) was much greater for TMA2 with 1.0 mm cores (Table 3, ‘Usable Cores’), the average score of the usable cores was not significantly different between the 0.6 and the 1.0 mm core TMAs (Table 3, ‘Core Quality’).
Although the quality of individual cores is the first step towards an effective TMA, the utility of the TMA is defined by the number of patients that can be analyzed after completion of the tissue stain. In this study, a patient’s sample was considered ‘analyzable’ if two or more cores had a usability score ≥2 (Figure 2). Even though the 1.0 mm core was more effective at capturing the target tissue (‘Usable Cores’, Table 2), the percentage of analyzable patients was not significantly different between the two TMAs (‘Analyzable Patients’, Table 3; tumor, P=1.000, normal, P=0.806, and CIS, P=0.286). This was simply because TMA1 (0.6 mm) contained one additional core for each tissue. This increase in the number of 0.6 mm cores compensated for the lower accuracy of each individual core.
Analytical Accuracy
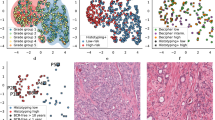
We defined the analytical accuracy by measuring the concordance between the donor tissue and the recipient cores and by calculating the inter-core variability between replicate cores taken from the same patient in the same TMA replicate. This was accomplished on sections from TMA1 and TMA2 stained for Ki67, collagen, and nuclei (Hoechst, Supplementary Figure 2). Fluorescent images of the stained cores and the donor-block whole sections were processed and the proliferation index was quantified as the percent Ki67-positive nuclei (Figure 3). Concordance was evaluated by plotting the Ki67-positive nuclei in the cores against the percent Ki67-positive nuclei in the whole section. The 1.0 mm core size (TMA2) demonstrated far greater donor-recipient correlation than the 0.6 mm cores (TMA1; Pearson correlation, TMA2, r=0.979, P=0.004 vs TMA1, r=0.478, P=0.415; Figure 4a).
Image processing. A digital RGB JPEG image of individual cores or a whole-slide section was processed and analyzed in ImageJ. (a) The RGB image is split into the individual R, G, and B channels. The individual gray-scale images are processed as follows: (i) ‘Thresholding’ to display the desired pixel intensity, (ii) ‘Fill Holes’ to fill empty spaces within non-uniform nuclei, (iii) ‘Watershed’ segmentation to separate partially overlapping nuclei. The ‘Analyze Particle’ function is subsequently applied to quantify the number of nuclei identified in each of the Red channel (Ki67+ nuclei) and Blue channel (All nuclei). (b) The ImageJ workflow.
Analytical accuracy. Analytical accuracy was evaluated by analyzing the proliferative status in the cores and correlating this to the proliferative status determined in the whole slide. (a) The mean % Ki67+ nuclei identified in the cores was graphed against the % Ki67+ identified in the whole-slide section. Individual data from five patients in each TMA is shown along with corresponding linear regression and 95% confidence interval. The quality of the correlation was assessed using Pearson correlation and two-tailed significance. (b) The mean % Ki67+ nuclei identified in the cores was graphed against average number of mitotic nuclei identified visually in 10 high-power fields (HPF) in the whole-slide section. Individual data from 10 patients in each TMA is shown along with corresponding linear regression and 95% confidence interval. The quality of the correlation was assessed using Pearson correlation and two-tailed significance.
In a separate analysis, we compared a manually scored mitotic index22 of the whole sections (donor) with percent Ki67-positive nuclei in the cores (Supplementary Table 1). Concordance was evaluated by plotting proliferation indices of the usable cores against the proliferation indices of the whole-slide section. The correlation between mitotic indices and percent Ki67-positive nuclei correlated strongly for the 1.0 mm core (TMA2), but not for the 0.6 mm core (TMA1) size (Pearson correlation, TMA2, r=0.669, P=0.035 vs TMA1, r=−0.200, P=0.579; Figure 4b).
Last, inter-core variability was evaluated by determining the standard deviation for replicate cores taken from the same tissue/donor block (Supplementary Table 1). The variability was consistently smaller for the 1.0 mm cores than the 0.6 mm cores. This reduced inter-core variability is important for improving the analytical prowess of the data as is evident by the superior 95% confidence interval of 1.0 mm cores (TMA2) in the correlation of percent Ki67-positive nuclei between donor whole sections and recipient cores (Figure 4). These observations were confirmed by staining for cytokeratin and assessing concordance between the cores and the whole section (Supplementary Figures 2 and 3).
Discussion
In TMA construction, there are two significant challenges that must be considered: (i) the cores must capture the target tissue (technical accuracy) and (ii) analysis of the tissue captured in a core must accurately correspond to a similar analysis of the full-size tissue it was taken from (analytical accuracy). This is particularly challenging for small histological features such as the thin layer of urothelium or CIS, as well as heterogeneous tissues such as nested squamous differentiation in urothelial carcinoma. Smaller core sizes can greatly increase the TMA density and limit depletion of the donor block (Figure 1). However, the smaller surface area of this core also reduces its representation of the original target tissue and its ability of capturing the desired histological feature.
We generated two pilot TMAs of 0.6 and 1.0 mm core size to compare and contrast the technical and analytical accuracy of these different core sizes in preparation of a large 2500 core TMA that will represent the urothelial carcinomas treated by cystectomy at Vanderbilt University Medical Center (VUMC) from 2000 to 2014. We hypothesized that the larger 1.0 mm core would have a better technical accuracy because the area of tissue it captures is nearly 3 × larger than that of the 0.6 mm core (Figures 1, 0.785 mm2 vs 0.2826 mm2). Although this was clearly true (‘Analyzable Cores’, Table 3), the lower technical accuracy of a 0.6 mm core was effectively compensated for by increasing the number of replicate cores/tissue/TMA from 2 to 3 (‘Analyzable Patients’, Table 3). Even at three replicates, a TMA of 0.6 mm core consumes less than half the amount of tissue used by a 1.0 mm core with two replicates (3 × 0.283 mm2=0.849 mm2 vs 2 × 1.0 mm2=2.0 mm2). However, unexpectedly, the 0.6 mm TMA (three cores/sample) was less analytically accurate than 1.0 mm TMA (two cores/sample; Figure 4). The lack of concordance for Ki67 staining was particularly striking because it demonstrated that even when a core captures the correct tissue, the staining analysis within 0.6 mm cores does not correspond well with analysis of the whole-slide section (Pearson correlation of 0.478, P=0.415, Figure 4a). The large inter-core variability of 0.6 mm cores and the corresponding broad 95% confidence interval suggests that variations due to tissue heterogeneity present within the donor block are amplified when small pieces are captured in 0.6 mm cores (Supplementary Table 1 and Figure 4, respectively). We speculate that in a heterogeneous tissue, one core may capture one extreme aspect of the tissue while another core captures the opposing extreme. This effect is more pronounced in smaller tissue sampling (eg, 0.6 mm core), ultimately leading to larger variability, which limits the analytical accuracy of such a TMA (Figure 4a and b). These observations suggest that there might be benefits to increasing the spot size beyond 1.0 mm as 1.5, 2, and even 4.0 mm cores can be generated. Although we did not directly test that assumption, we speculate little change in the technical and analytic accuracy because the 1.0 mm cores already provide good concordance with the intact tissue. Moreover, the rapid loss of TMA density (Table 1) for cores >1.0 mm would diminish the benefit of mapping multiple patients into a single slide.
Although TMA construction has become a well-accepted practice for accomplishing high-volume parallel analysis of tissue from many specimens, very few studies have investigated the influence of parameters like core size with respect to the final quality and usability of the TMA. It is evident that capturing the desired tissue requires that the core punch be placed in the appropriate part of the block.5, 7, 23 Fonseca et al10 explored the contribution of core size to the quality of a TMA using measures similar to our ‘Technical Accuracy’ and concluded that it can be difficult to capture small features with a small core size. Increasing the number of cores is desirable but quickly increases the labor and cost of TMA production. To minimize these challenges, we used a next-generation TMA approach with digitized slides and an automated arrayer (3D Histotech,4 Supplementary Figure 1). Not only did this improve placement of the cores, the automated workflow facilitated the production of replicate TMA blocks (three replicate blocks for each TMA, Figure 1 and Supplementary Figure 1) with limited additional human labor.
Beyond TMA construction, the utility of the TMA depends largely on how accurately the staining of individual cores corresponds with staining of original donor tissue. In recent years, the staining procedures and subsequent analyses have become increasingly sophisticated. Single-color chromogenic stains are still common but multi-color fluorescent stains are increasingly necessary to determine correlations in expression and distribution between distinct molecular antigens.24 Consequently, the analysis of TMAs is transitioning from an ‘absent vs present’ or ordinal 1–4 scoring mechanism25 to fully computational analyses that make use of continuous signal intensity measures, integration of surface area and maximal signal intensity, signal co-localization, and even single-cell segmentation.26 Our assessment of ‘analytical accuracy’ takes these computational evaluations into consideration and revealed a striking advantage of using a larger (1.0 mm) core size when considering concordance between the cores and their donor block as well as the variability between replicate cores.
Conclusion
In TMA construction, a 1.0 mm core provides both technical and analytical advantages over a 0.6 mm core size. These benefits outweigh the increased replicate sampling and TMA patient density gained with 0.6 mm cores. Based on these findings, we have constructed a large urothelial carcinoma TMA using 1.0 mm cores and advise others to consider both technical and analytical accuracy when constructing their own TMAs.
References
Battifora H . The multitumor (sausage) tissue block: novel method for immunohistochemical antibody testing. Lab Invest 1986; 55: 244–248.
Kononen J, Bubendorf L, Kallionimeni A et al, Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med 1998; 4: 844.
Richter J, Wagner U, Kononen J et al, High-throughput tissue microarray analysis of cyclin E gene amplification and overexpression in urinary bladder cancer. Am J Pathol 2000; 157: 787–794.
Gustavson MD, Rimm DL, Dolled-Filhart M . Tissue microarrays: leaping the gap between research and clinical adoption. Pers Med 2013; 10: 441.
Zlobec I, Koelzer VH, Dawson H et al, Next-generation tissue microarray (ngTMA) increases the quality of biomarker studies: an example using CD3, CD8, and CD45RO in the tumor microenvironment of six different solid tumor types. J Transl Med 2013; 11: 104.
Nocito A, Bubendorf L, Tinner EM et al, Microarrays of bladder cancer tissue are highly representative of proliferation index and histological grade. J Pathol 2001; 194: 349–357.
Voduc D, Kenney C . Tissue microarrays in clinical oncology. Semin Radiat Oncol 2008; 18: 89.
Iakovlev VV, Pintilie M, Morrison A et al, Effect of distributional heterogeneity on the analysis of tumor hypoxia based on carbonic anhydrase IX. Lab Invest 2007; 87: 1206–1217.
Jones S . Comparative evaluation of high-throughput small-core (0.6-mm) and large-core (2-mm) thyroid tissue microarray: is larger better? Arch Pathol Lab Med 2012; 136: 199.
Fonseca FP, de Andrade BAB, Rangel ALCA et al, Tissue microarray is a reliable method for immunohistochemical analysis of pleomorphic adenoma. Oral Surg Oral Med Oral Pathol Oral Radiol 2013; 117: 81–88.
Remotti H . Tissue microarrays: construction and use. Methods Mol Biol 2013; 980: 13–28.
Quintayo MA, Starczynski J, Yan FJ et al, Virtual tissue microarrays: a novel and viable approach to optimizing tissue microarrays for biomarker research applied to ductal carcinoma in situ. Histopathology 2014; 65: 2–8.
Rubin MA, Dunn R, Strawderman M et al, Tissue microarray sampling strategy for prostate cancer biomarker analysis. Am J Surg Pathol 2002; 26: 312–319.
Wagener N, Bulkescher J, Macher-Goeppinger S et al, Endogenous BTG2 expression stimulates migration of bladder cancer cells and correlates with poor clinical prognosis for bladder cancer patients. Br J Cancer 2013; 108: 973.
Munari E, Chaux A, Maldonado L et al, Cyclin A1 expression predicts progression in pT1 urothelial carcinoma of bladder: a tissue microarray study of 149 patients treated by transurethral resection. Histopathology 2015; 66: 262.
Wang H, Albadine R, Magheli A et al, Increased EZH2 protein expression is associated with invasive urothelial carcinoma of the bladder. Urol Oncol 2012; 30: 428.
Kaufman DS, Shipley WU, Feldman AS . Bladder cancer. Lancet 2009; 374: 239–249.
Harris P A, Taylor R, Thielke R et al, Research electronic data capture (REDCap) - A metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform 2009; 42: 377–381.
Fedor HL, De Marzo AM . Practical methods for tissue microarray construction. Methods Mol Med 2004; 103: 89–101.
Krahn KN, Bouten CVC, van Tuijl S et al, Fluorescently labeled collagen binding proteins allow specific visualization of collagen in tissues and live cell culture. Anal Biochem 2006; 350: 177–185.
Hartig SM . Basic image analysis and manipulation in ImageJ. Curr Protoc Mol Biol 2013; 14: 14.15.
Goyal S, Singh UR, Sharma S et al, Correlation of mitotic indices, AgNor count, Ki-67 and Bcl-2 with grade and stage in papillary urothelial bladder cancer. Urol J 2014; 11: 1238–1247.
Kyndi M, Sørensen FB, Knudsen H et al, Tissue microarrays compared with whole sections and biochemical analyses. A subgroup analysis of DBCG 82 b&c. Acta Oncol 2008; 47: 591.
Tam J, Cordier GA, Borbely JS et al, Cross-talk-free multi-color STORM imaging using a single fluorophore. PLoS One 2014; 9: e101772.
Gavrielides MA, Conway C, O'Flaherty N et al. Observer performance in the use of digital and optical microscopy for the interpretation of tissue-based biomarkers. Anal Cell Pathol (Amst) 2014; 2014: 157308.
Amaral T, McKenna SJ, Robertson K et al, Classification and immunohistochemical scoring of breast tissue microarray spots. IEEE Trans Biomed Eng 2013; 60: 2806–2814.
Acknowledgements
This work was supported by the Canadian Collaborative Research & Innovation Opportunities (CRIO) to AZ and EH, Patients Centered Outcome Research Initiative (PCORI)—Pathlink pilot funding to AZ, and the RedCap development effort is maintained with support from the NIH (UL1 TR000445 from NCATS/NIH). SAAE was supported by VA CDA IK2BX002498. Support was also provided through the Vanderbilt Ingram Cancer Center support grant.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Laboratory Investigation website
Tissue-microarrays are critical for simultaneous analysis of large numbers of samples and patients. Cores in a tissue-microarray must accurately represent the target tissue in order to provide an effective research tool. This paper demonstrates that core size significantly impacts accuracy, where 1.0 mm cores show superior technical and analytical accuracy over smaller 0.6 mm cores.
Rights and permissions
About this article
Cite this article
Eskaros, A., Egloff, S., Boyd, K. et al. Larger core size has superior technical and analytical accuracy in bladder tissue microarray. Lab Invest 97, 335–342 (2017). https://doi.org/10.1038/labinvest.2016.151
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/labinvest.2016.151
This article is cited by
-
PAM staining intensity of primary neuroendocrine neoplasms is a potential prognostic biomarker
Scientific Reports (2020)