Abstract
Benign fibrous histiocytomas (FH) can be subdivided into several morphological and clinical subgroups. Recently, gene fusions involving either one of two protein kinase C genes (PRKCB and PRKCD) or the ALK gene were described in FH. We here wanted to evaluate the frequency of PRKCB and PRKCD gene fusions in FH. Using interphase fluorescence in situ hybridization on sections from formalin-fixed paraffin-embedded (FFPE) tumors, 36 cases could be analyzed. PRKCB or PRKCD rearrangements were seen in five tumors: 1/7 regular, 0/3 aneurysmal, 0/6 cellular, 2/7 epithelioid, 0/1 atypical, 2/10 deep, and 0/2 metastatic lesions. We also evaluated the status of the ALK gene in selected cases, finding rearrangements in 3/7 epithelioid and 0/1 atypical lesions. To assess the gene fusion status of FH further, deep sequencing of RNA (RNA-Seq) was performed on FFPE tissue from eight cases with unknown gene fusion status, as well as on two FH and six soft tissue sarcomas with known gene fusions; of the latter eight positive controls, the expected fusion transcript was found in all but one, while 2/8 FH with unknown genetic status showed fusion transcripts, including a novel KIRREL/PRKCA chimera. Thus, also a third member of the PRKC family is involved in FH tumorigenesis. We conclude that gene fusions involving PRKC genes occur in several morphological (regular, cellular, aneurysmal, epithelioid) and clinical (cutaneous, deep) subsets of FH, but they seem to account for only a minority of the cases. In epithelioid lesions, however, rearrangements of PRKC or ALK were seen, as mutually exclusive events, in the majority (5/7) of cases. Finally, the study also shows that RNA-Seq is a promising tool for identifying gene fusions in FFPE tissues.
Similar content being viewed by others
Main
Benign fibrous histiocytoma (FH) can be subdivided into several morphological and clinical subgroups. Morphological variants include cellular, aneurysmal, epithelioid, and atypical types, and clinical manifestations range from benign tumors of the skin, deep soft tissues or skeleton to, rarely, metastasizing tumors.1, 2, 3 We recently showed that both cutaneous and deep FH may carry specific gene fusions, in which a member—either PRKCB and PRKCD—of the gene family encoding protein kinase C (PRKC) is juxtaposed with a gene encoding a membrane-associated protein.4 The pathogenetic mechanism was assumed to involve the uncoupling of the carboxy-terminal kinase domain of the PRKC protein from its regulatory domain, and its ectopic localization through fusion with the amino-terminal part of a membrane-associated protein. It has also recently been shown that some cases of epithelioid, and possibly also atypical, FH may harbor fusions activating the ALK protein.5, 6 The connection between ALK rearrangements and FH was further evaluated and strengthened by Doyle et al., showing that ALK fusions were restricted to the epithelioid subtype.7
To study the frequency and distribution of PRKCB and PRKCD fusions in FH, we used interphase fluorescence in situ hybridization (FISH) on a series of tumors that represented different morphological and clinical subsets; the results of that study prompted transcriptome sequencing (RNA-Seq) of RNA from formalin-fixed paraffin-embedded (FFPE) tissue from a second set of FH.
Materials and Methods
Patients and Tumors
For the interphase FISH studies, 66 cases of FH were retrieved from the consultation files of one of the authors (CDM Fletcher). The cases were selected to represent different clinical and morphological aspects of FH: 10 regular, 10 cellular, 10 aneurysmal, 10 atypical, 10 epithelioid, 11 deep, and 5 metastatic lesions; no prior genetic information was available on these tumors, except for two epithelioid FH that had been analyzed with regard to ALK gene rearrangements. The diagnostic criteria have been outlined before.1, 8, 9, 10, 11, 12, 13 Two cases of regular FH with known fusions involving PRKCB and PRKCD (previously published as Cases 5 and 6 in4), respectively, were used as positive controls. All seven epithelioid FH that could be analyzed by FISH were also analyzed with regard to expression of the ALK protein, as described.7
For the RNA-Seq study, another set of six recently (2011–2013) diagnosed cutaneous regular FH was chosen. In addition, we included two cytogenetically analyzed FH from which no frozen tissue was available: one cellular/aneurysmal FH from 1990 (Case 7 in Plaszczyca et al.4 and Case Lu 317 in Vanni et al.14) with an unbalanced t(3;12) and one cellular FH from 2010 with a balanced t(1;16)(p36;p11), strongly suggestive of a PDPN/PRKCB fusion. As positive controls, we selected two FH from 2009 with PDPN/PRKCB fusions (Cases 2 and 5 in Plaszczyca et al.4), four myxoid liposarcomas (MLS) from 2011 or 2014 with FUS/DDIT3 fusions, one synovial sarcoma from 2013 with an SS18/SSX2 fusion, and one low-grade fibromyxoid sarcoma from 2013 with an FUS/CREB3L2 fusion; the fusion gene status of the positive controls had been established by G-banding, FISH, and/or RT-PCR (data not shown). Thus, the RNA-Seq study included eight FH with unknown fusion gene status and eight soft tissue tumors, including two cases of FH, with known gene fusions.
FISH
Interphase FISH for the detection of rearrangements of PRKCB and PRKCD, mapping to chromosome bands 16p12 and 3p21, respectively, was performed on 4 μm FFPE tumor sections. The bacterial artificial chromosome (BAC) probes were obtained from the BACPAC Resource Center (http://bacpac.chori.org; Supplementary Table 1). Clone preparation, hybridization, and analysis were performed as described.15, 16 On average 113 (range=83–176) nuclei from different areas were evaluated per probe set in each case. Nuclei displaying separate green and red signals or a single 3′ signal were scored as positive. In seven epithelioid and one atypical FH, the status of the ALK gene was also investigated, using a commercial double color, break-apart probe (LSI ALK, Vysis, Abbott Molecular, IL, USA); in each case, 95–112 nuclei were scored. The status of the PRKCA gene was evaluated in Case 39, using probes described in Supplementary Table 1; 93 nuclei were scored.
RNA-Seq and RT-PCR
For RNA extraction, tumor blocks from each of the 16 cases (Cases 37–44 and PC3–10) were retrieved and tumor-representative areas were selected. Then, 10 μm sections were cut from each tumor (n=3–38, depending on the size of the selected areas; the aim was to achieve a total of 450 mm2 of tumor tissue from each case). The first two sections were discarded and the following sections were immediately put in Deparaffinization Solution (Qiagen, Valencia, CA, USA) followed by RNA extraction with Qiagen’s RNeasy FFPE Kit, according to the manufacturer’s recommendations. The quality and quantity of the RNA, with a particular focus on the fraction of RNA fragments >200 nt (the DV200), were measured using an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA). mRNA libraries were prepared from 20 to 50 ng, depending on the DV200 value, of the extracted RNA using the capturing chemistry of the TruSeq RNA Access Library Prep Kit (Illumina, San Diego, CA, USA), according to the manufacturer’s recommendations. The quality of the mRNA library was measured using an Agilent 2100 Bioanalyzer (Agilent Technologies) and the concentration was measured on a Qubit Fluorometer using the Qubit RNA HS Assay kit (Life Technologies, Rockville, MD, USA). With a loading concentration of 1.8 pM, paired-end 2 × 81 nt reads were generated in-house from the mRNA libraries using Illumina’s High Output Kit (150 cycles) on a NextSeq 500 (Illumina). To identify candidate fusion transcripts from the sequence data, analyses were performed on fastq files using ChimeraScan version 0.4.5 and TopHat version 2.0.7,17, 18 as decribed.19, 20 The GRCh37/hg19 build was used as the human reference genome.
RT-PCR was performed in Case 39, using primers specific for KIRREL (forward, 5'-GACCGGGAGGATGACACCG-3') and PRKCA (reverse, 5'-GGGACTGATGACTTTGTTGCC-3'). RT-PCR and sequencing were performed as described.16
Results
FISH
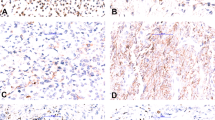
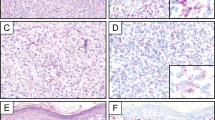
For technical reasons, interphase FISH for PRKCB and/or PRKCD rearrangements was possible to perform in only 36 of the selected FH cases (cases 1–36) and in two positive controls (PC1–2; Table 1); the remaining samples did not yield signals of sufficient quality. The distribution of positive nuclei differed between the two probe sets: the mean numbers of positive nuclei for PRKCB and PRKCD rearrangements were 8.4% and 15.5%, with standard deviations (s.d.) 7.1% and 11.3%, respectively. Cutoff values for scoring cases as positive were chosen at >23% for PRKCB and >38% for PRKCD (i.e., mean+2 s.d.). The cutoff for the commercial ALK probe was set at >15%. Using these criteria, the two positive controls, i.e., tumors in which we previously had identified fusions using RT-PCR on frozen tissue,4 were positive for the expected PRKC gene and negative for the other (Table 1). PRKCB was scored as positive in 3/34 cases and PRKCD was positive in 2/31 cases. The PRKC-positive FH cases were distributed as follows: 1/7 regular, 0/3 aneurysmal, 0/6 cellular, 2/7 epithelioid, 0/1 atypical, 2/10 deep, 0/2 metastatic (Table 1). The two epithelioid FH that had PRKCB rearrangements at FISH analysis were negative for ALK protein expression (Table 1).
The status of the ALK gene had previously been studied in two cases of epithelioid FH, one being negative and one positive.7 Here, these results could be repeated, and another two cases of epithelioid FH, both negative for PRKC rearrangements, were scored as positive. Thus, 3/7 epithelioid and 0/1 atypical FH were ALK positive (Table 1).
The status of the PRKCA gene was investigated by interphase FISH on cut sections from Case 39, in which a KIRREL/PRKCA fusion transcript was found by both RNA-Seq and RT-PCR (described below). The FISH analysis disclosed a split signal, consistent with translocation, in 42% of the nuclei.
RNA-Seq and RT-PCR
RNA of sufficient quantity (range=2.9–43.4 μg, median=9.7 μg) and quality (DV200 value: range=45–75%, mean=60%) could be extracted from all 16 tumors. The number of reads obtained per case ranged from 14.5 to 64.4 × 106, median=25.2 × 106 (Table 2). The two software used in this study identified numerous potential fusion transcripts in all cases (Supplementary Tables 2 and 3), the vast majority of which could be discarded as read-through transcripts or technical artifacts. A single, biologically relevant fusion transcript, all of which were identified by ChimeraScan, was found in nine cases; five of these were found also with TopHat (Table 2). The expected fusion transcripts were found in 5/6 sarcomas with known gene fusions (three MLS with the FUS/DDIT3 fusion, one synovial sarcoma with the SS18/SSX2 fusion, and one low-grade fibromyxoid sarcoma with the FUS/CREB3L2 fusion); one MLS did not display the expected FUS/DDIT3 fusion by any of the two software. Also the two FH with known PDPN/PRKCB fusions were identified at RNA-Seq of FFPE tissue. Thus, 7/8 positive controls were correctly classified. In addition, one FH with a known t(1;16) showed a PDPN/PRKCB fusion, in agreement with the karyotype, whereas one FH with an unbalanced t(3;12) was negative. Of the remaining six FH (Cases 37–44), five did not display any relevant fusion transcript, whereas one showed a novel KIRREL/PRKCA fusion (Table 2). This fusion was verified by RT-PCR, revealing an in-frame fusion of exon 13 (nt 158,093,762) of KIRREL with exon 9 (nt 66,732,687) of PRKCA (Positions according to GRCh38; Figure 1).
Discussion
We recently showed that gene fusions involving the PRKCB and PRKCD genes are recurrent in FH.4 However, that study was biased in the sense that it only included tumors that had clonal chromosomal rearrangements at G-banding analysis, potentially enriching certain genetic subgroups with a high proliferative capacity in vitro. Furthermore, only four cases in that study could be analyzed by molecular and/or FISH techniques. We thus wanted to explore a larger series of BFH, with tumors representing all known morphologic subtypes. For that purpose, unstained sections from 66 cases of BFH were retrieved and studied with regard to PRKCB and PRKCD rearrangements using interphase FISH. However, this analysis turned out to be unusually challenging in the sense that it was very difficult to obtain signals of sufficient quality. Because FISH analyses with commercial probes for the ALK gene have been successfully performed on FFPE sections from FH,7 we assume that the technical problems were due to the probe sets we used. However, also the age of the samples could possibly have affected the results. Nevertheless, reliable results could be obtained in 36 of the cases; 5 of them were interpreted to have a PRKCB or a PRKCD rearrangement (Table 1). As this frequency (5/36, 14%) of PRKCB/PRKCD-positive cases was considerably lower than in our initial study and because the positive cases were found among several of the different morphologic/clinical subsets—regular, epithelioid, and deep lesions—we wanted to use another technique to investigate whether the discrepancy was due to poor sensitivity of FISH. With no further material being available from the intial cohort, a second set of recently diagnosed regular FH was retrieved for RNA extraction and RNA-Seq. This approach turned out to be technically highly successful, in terms of RNA quantity and quality as well as sequencing results (Table 2). With only 1/8 positive controls being negative for the expected gene fusion, it seems reasonable to assume that the results on the FH with unknown genetic status were fairly accurate; the negative case (PC9) was the positive control yielding the lowest number of reads, possibly explaining the negative results (Table 2).
The RNA-Seq analysis identified one cellular FH with the PDPN/PRKCB fusion that has been previously detected in FH.4 In addition, 1/6 regular FH displayed a novel fusion between KIRREL and PRKCA, mapping to 1q23 and 17q24, respectively. Although KIRREL has not been implicated in gene fusions before, Bridge and co-workers recently described a recurrent SLC44A1/PRKCA fusion in papillary glioneural tumor, with the same breakpoint in PRKCA as in the FH of the present study.21 Further arguments for a pathogenetic significance of the KIRREL/PRKCA fusion in the context of FH is that KIRREL, like the previously identified amino-terminal partners PDPN, CD63, and LAMTOR1, is a membrane-associated protein, retaining its membrane-binding part in the chimeric protein.4
We can thus conclude that PRKC gene rearrangements are indeed recurrent in FH and that, based on the present and our previous study, they at least occur in the regular, cellular, aneurysmal, and epithelioid subtypes, and that they may be found in both cutaneous and deep lesions; more cases of atypical and metastatic FH need to be analyzed before it can be excluded that also these entities may harbor PRKC fusions. The overall frequency of PRKC-positive FHs can only be roughly estimated. First, interphase FISH was only performed for two members of the PRKC gene family (PRKCB and PRKCD) but we now know from the RNA-Seq study that at least the alpha variant (PRKCA) can also be involved in fusions. Second, results of interphase FISH on FFPE sections are somewhat difficult to standardize, as the samples may vary significantly in technical quality. If we had lowered the cutoff values to 1 s.d. above the mean frequencies of split signals, four more cases would have been scored as positive for PRKC rearrangements, whereas an increase of the cutoff to 3 s.d. above mean frequencies would have turned two PRKCD-positive cases, including the positive control, and two PRKCB-positive cases into negative ones (Table 1); unfortunately, no tissue blocks for RNA extraction were available from the cases that were used for FISH. Taking the methodological shortcomings mentioned above into account, our results from FISH and RNA-Seq suggest that only some 15–25% of FH are positive for PRKC gene fusions. Thus, there must exist alternative pathogenetic pathways in FH. Indeed, it was recently shown by both FISH and RNA-Seq that epithelioid FH carry fusions involving the ALK gene—VCL/ALK and SQSTM1/ALK, respectively.6 This finding was further expanded by Doyle and co-workers who analyzed 33 epithelioid FH by immunohistochemistry (IHC)—29/33 cases were ALK-positive, and 14/14 of these turned out to be ALK-rearranged also at FISH analysis; other subtypes of FH were IHC-negative for ALK.7 Interestingly, epithelioid FH was the subtype most often (2/7) showing a PRKC gene rearrangement in our FISH study (Table 1), suggesting that the vast majority of these lesions harbor either an ALK fusion or a PRKC fusion. The conclusion that ALK and PRKC activation constitute alternative, and not overlapping, pathogenetic pathways in epithelioid FH was supported by the finding that the two cases with PRKCB rearrangement were negative for ALK expression, whereas the other five cases were positive (Table 1); there were no obvious morphological or clinical differences between PRKC- and ALK-positive epithelioid FH, but it should be kept in mind that very few cases were analyzed. Also two tumors diagnosed as atypical FH were recently reported to have ALK rearrangements, as shown by FISH or IHC,5 but, in our opinion, the morphologic description and images would fit better with epithelioid FH. The single atypical FH (Case 24) that we could analyze by FISH was negative for PRKCB, PRKCD, and ALK rearrangements (Table 1).
The RNA-Seq part of our study adds to the still small but growing number of reports showing that the NGS technologies are applicable to RNA extracted from FFPE tissues.22, 23, 24, 25, 26 Furthermore, RNA-Seq on FFPE tissue has only rarely been used to detect gene fusions.6, 23, 27 The study of Sweeney and co-workers is of particular interest in the present context, as they also investigated sarcomas (three synovial sarcomas, three MLS, two Ewing sarcomas, and one clear cell sarcoma), used Illumina equipment and kits, and had results similar to ours;23 all their cases showed the expected gene fusions, although some of the samples had to be re-analyzed at greater depth or after depletion of ribosomal RNA before obtaining positive results. Some of the methodological differences between their and our study deserve further comments. First, Sweeney and co-workers read longer sequences than we did (2 × 150 nt, compared with 2 × 81 nt in our study). In principle, longer reads should have a greater chance of covering the fusion breakpoints (spanning reads) and while all their cases had fusion spanning reads, three of our positive controls did not (Table 2). As shown in our study, however, flanking reads can also accurately identify gene fusions. Second, none of the samples analyzed by Sweeney and co-workers had been archived for more than 6 months, whereas our oldest positive sample had been archived for 5 years. Most likely, even older samples can be successfully studied with regard to gene fusion status. Hedegaard et al. performed a comprehensive analysis of RNA-Seq data on both FFPE and fresh frozen samples from various carcinomas that had been stored up to 244 months; the two methods showed very good correspondence in terms of up- and downregulated genes.24 Third, Sweeney and co-workers selected reads aligning only to genes known to be involved in sarcoma-associated fusions, a so-called sarcomatome, whereas we extracted all potential fusion genes. Our strategy did not result in any potential false positives, and did not significantly increase the time needed to identify the fusion of interest. Most importantly, our unbiased approach allowed us to find a novel fusion transcript, where none of the two genes had been implicated in soft tissue tumors before. In summary, we find the results of our RNA-Seq study very promising and believe that FFPE samples can be successfully used for gene fusion detection.
References
Gleason BC, Fletcher CD . Deep “benign” fibrous histiocytoma: clinicopathologic analysis of 69 cases of a rare tumor indicating occasional metastatic potential. Am J Surg Pathol 2008;32:354–362.
Luzar B, Calonje E . Cutaneous fibrohistiocytic tumours—an update. Histopathology 2010;56:148–165.
Nielsen GP, Kyriakos M. Non-ossifying fibroma/benign fibrous histiocytoma of bone. In: Fletcher CD, Bridge JA, Hogendoorn PCW, Mertens F (eds). World Health Organization Classification of Tumours. Pathology and Genetics of Tumours of Soft Tissue and Bone. IARC Press: Lyon, 2013, pp 302–304.
Plaszczyca A, Nilsson J, Magnusson L et al. Fusions involving protein kinase C and membrane-associated proteins in benign fibrous histiocytoma. Int J Biochem Cell Biol. 2014;53:475–481.
Szablewski V, Laurent-Roussel S, Rethers L et al. Atypical fibrous histiocytoma of the skin with CD30 and p80/ALK1 positivity and ALK gene rearrangement. J Cutan Pathol 2014;41:715–719.
Jedrych J, Nikiforova M, Kennedy TF et al. Epithelioid cell histiocytoma of the skin with clonal ALK gene rearrangement resulting in VCL-ALK and SQSTM1-ALK gene fusions. Br J Dermatol 2015;172:1427–1429.
Doyle LA, Marino-Enriquez A, Fletcher CD et al. ALK rearrangement and over-expression in epithelioid fibrous histiocytoma. Mod Pathol, in press.
Calonje E, Mentzel T, Fletcher CD . Cellular benign fibrous histiocytoma. Clinicopathologic analysis of a distinctive variant of cutaneous fibrous histiocytoma with frequent recurrence. Am J Surg Pathol 1994;18:668–676.
Singh Gomez C, Calonje E, Fletcher CD . Epithelioid benign fibrous histiocytoma of skin: clinic-pathological analysis of 20 cases of a poorly known variant. Histopathology 1994;24:123–129.
Calonje E, Fletcher CD . Aneurysmal benign fibrous histiocytoma: clinicopathological analysis of 40 cases of a tumour frequently misdiagnosed as a vascular neoplasm. Histopathology 1995;26:323–331.
Kaddu S, McMenamin M, Fletcher CD . Atypical fibrous histiocytoma of the skin. Clinicopathologic analysis of 59 cases with evidence of infrequent metastasis. Am J Surg Pathol 2002;26:35–46.
Doyle LA, Fletcher CD . EMA positivity in epithelioid fibrous histiocytoma: a potential diagnostic pitfall. J Cutan Pathol 2011;38:697–703.
Doyle LA, Fletcher CD . Metastasizing “benign” cutaneous fibrous histiocytoma. A clinicopathologic analysis of 16 cases. Am J Surg Pathol 2013;37:484–495.
Vanni R, Fletcher CD, Sciot R et al. Cytogenetic evidence of clonality in cutaneous benign fibrous histiocytomas: A report of the CHAMP study group. Histopathology 2000;37:212–217.
Arbajian E, Puls F, Magnusson L et al. Recurrent EWSR1-CREB3L1 gene fusions in sclerosing epithelioid fibrosarcoma. Am J Surg Pathol 2014;38:801–808.
Walther C, Tayebwa J, Lilljebjörn H et al. A novel SERPINE1-FOSB fusion results in transcriptional up-regulation of FOSB in pseudomyogenic hemangioendothelioma. J Pathol 2014;232:534–540.
Iyer MK, Chinnaiyan AM, Maher CA . ChimeraScan: a tool for identifying chimeric transcription in sequencing data. Bioinformatics 2011;27:2903–2904.
Kim D, Pertea G, Trapnell C et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 2013;14:R36.
Hofvander J, Tayebwa J, Nilsson J et al. Recurrent PRDM10 gene fusions in undifferentiated pleomorphic sarcoma. Clin Cancer Res 2015;21:864–869.
Hofvander J, Tayebwa J, Nilsson J et al. RNA sequencing of sarcomas with simple karyotypes: identification and enrichment of fusion transcripts. Lab Invest 2015;95:603–609.
Bridge JA, Liu X-Q, Sumegi J et al. Identification of a novel, recurrent SLC44A1-PRKCA fusion in papillary glioneuronal tumor. Brain Pathol 2013;23:121–128.
Adiconis X, Borges-Rivera D, Satija R et al. Comparative analysis of RNA sequencing methods for degraded or low-input samples. Nat Methods 2013;10:623–629.
Sweeney RT, Zhang B, Zhu SX et al. Desktop transcriptome sequencing from archival tissue to identify clinically relevant translocations. Am J Surg Pathol 2013;37:796–803.
Hedegaard J, Thorsen K, Lund MK et al. Next-generation sequencing of RNA and DNA isolated from paired fresh-frozen and formalin-fixed paraffin-embedded samples of human cancer and normal tissue. PLoS ONE 2014;9:e98187.
Li P, Conley A, Zhang H, Kim HL . Whole-transcriptome profiling of formalin-fixed, paraffin-embedded renal cell carcinoma by RNA-seq. BMC Genomics 2014;15:1087.
Zhao W, He X, Hoadley KA et al. Comparison of RNA-Seq by poly (A) capture, ribosomal RNA depletion, and DNA microarray for expression profiling. BMC Genomics 2014;15:419.
Ma Y, Ambannavar R, Stephans J et al. Fusion transcript discovery in formalin-fixed paraffin-embedded human breast cancer tissues reveals a link to tumor progression. PLoS ONE 2014;9:e94202.
Acknowledgements
We thank Caroline Jansson for technical assistance with the cutting of FFPE sections for the RNA-Seq studies and Henrik Lilljebjörn for valuable discussions. This study was supported by the Swedish Cancer Society and the Gunnar Nilsson Cancer Foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Laboratory Investigation website
The frequency and distribution of gene fusions involving protein kinase C (PRKC) were studied in the many different morphological and clinical subsets of benign fibrous histiocytoma. Using FISH and RNA sequencing on formalin-fixed, paraffin-embedded samples, such fusions were detected in most subtypes. In epithelioid fibrous histiocytoma, PRKC and ALK rearrangements were mutually exclusive.
Rights and permissions
About this article
Cite this article
Walther, C., Hofvander, J., Nilsson, J. et al. Gene fusion detection in formalin-fixed paraffin-embedded benign fibrous histiocytomas using fluorescence in situ hybridization and RNA sequencing. Lab Invest 95, 1071–1076 (2015). https://doi.org/10.1038/labinvest.2015.83
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/labinvest.2015.83
This article is cited by
-
Epithelioid fibrous histiocytoma: three diagnostically challenging cases with novel ALK gene fusions, unusual storiform growth pattern, and a prominent spindled morphology
Virchows Archiv (2022)
-
Broadening the spectrum of NTRK rearranged mesenchymal tumors and usefulness of pan-TRK immunohistochemistry for identification of NTRK fusions
Modern Pathology (2021)
-
ALK alterations in salivary gland carcinomas
Virchows Archiv (2021)
-
Cutaneous soft tissue tumors: how do we make sense of fibrous and “fibrohistiocytic” tumors with confusing names and similar appearances?
Modern Pathology (2020)
-
Aberrant receptor tyrosine kinase signaling in lipofibromatosis: a clinicopathological and molecular genetic study of 20 cases
Modern Pathology (2019)