Abstract
The dyslipidemia associated with type 2 diabetes mellitus (T2DM) is an important risk factor for atherosclerotic cardiovascular disease. However, until now little attention has been paid to the role that the intestine might have. The aim of this research was to determine the relation between insulin resistance and intestinal de novo lipogenesis/lipoprotein synthesis in morbidly obese subjects and to study the effect of insulin on these processes. Jejunal mRNA expression of the different genes involved in the intestinal de novo lipogenesis/lipoprotein synthesis was analyzed in three groups of morbidly obese subjects: Group 1 with low insulin resistance (MO-low-IR), group 2 with high insulin resistance (MO-high-IR), and group 3 with T2DM and treatment with metformin (MO-metf-T2DM). In addition, intestinal epithelial cells (IECs) from MO-low-IR were incubated with different doses of insulin/glucose. In Group 2 (MO-high-IR), the jejunal mRNA expression levels of apo A-IV, ATP-citrate lyase (ACLY), pyruvate dehydrogenase (lipoamide) beta (PDHB), and sterol regulatory element-binding protein-1c (SREBP-1c) were significantly higher and acetyl-CoA carboxylase alpha (ACC1) and fatty-acid synthase lower than in Group 1 (MO-low-IR). In Group 3 (MO-metf-T2DM), only the ACLY and PDHB mRNA expressions were significantly higher than in Group 1 (MO-low-IR). The mRNA expression of most of the genes studied was significantly linked to insulin and glucose levels. The incubation of IEC with different doses of insulin and glucose produced a higher expression of diacylglycerol acyltransferase 2, microsomal triglyceride transfer protein, apo A-IV, SREBP-1c, and ACC1 when both, glucose and insulin, were at a high concentration. However, with only high insulin levels, there were higher apo A-IV, PDHB and SREBP-1c expressions, and a lower ACLY expression. In conclusion, the jejunum of MO-high-IR has a decreased mRNA expression of genes involved in de novo fatty-acid synthesis and an increase of genes involved in acetyl-CoA and lipoprotein synthesis. This effect is attenuated by metformin. In addition, the expression of most of the genes studied was found to be regulated by insulin.
Similar content being viewed by others

Main
The dyslipidemia associated with obesity, insulin resistance and type 2 diabetes mellitus (T2DM) is an important risk factor for atherosclerotic cardiovascular disease. Previously, a retarded triglyceride-rich lipoproteins catabolism had been related with the development of these pathologies.1 Moreover, it is known that a high-fat diet can modify the composition of triglyceride-rich lipoprotein.2 However, little attention was paid to the role that the intestine could have in these metabolic diseases.
Recent studies point to the possibility that fasting overproduction of intestinal lipoproteins might also contribute to the dyslipidemia.3 There is a growing body of evidence that in the fasting state, the intestine synthesizes small triglyceride-rich lipoprotein, maintaining a basal production of apo B-48.4 Also, it has been shown that elevated fasting chylomicron levels are present in obese and hyperlipidemic subjects.5 Thus, the jejunum appears to have an active role in lipid homeostasis in the fasting state. We have previously shown that morbidly obese subjects with type 2 diabetes had lower jejunal lipid content in fasting status than non-diabetic morbidly obese subjects, and that this correlated negatively with apolipoprotein A-IV (apo A-IV) mRNA expression.3 Apo A-IV has an important role in enhancing the secretion of chylomicrons and associated lipids in newborn enterocytes.6
A dysregulation of intestinal lipoprotein metabolism in insulin-resistant states has also been shown in animal models7 and humans.8 Insulin resistance and T2DM might enhance intestinal lipoprotein production, involving increased de novo lipogenesis and chylomicron production. De novo lipogenesis is controlled by several transcription factors. In general, the lipogenic gene expression is regulated by the sterol regulatory element-binding protein-1c (SREBP-1c), which upregulates enzymes involved in fatty-acid synthesis (acetyl-CoA carboxylase alpha (ACC1) and fatty-acid synthase (FAS)9), as well as enzymes that supply acetyl-CoA, (ATP-citrate lyase (ACLY)). On the other hand, SREBP-1c is induced by insulin, accounting for the lipogenic effect of chronic hyperinsulinemia.9 In insulin-resistant animal models, increased SREBP-1c has been demonstrated in enterocytes and might explain the elevation of de novo lipogenesis and the increased intracellular pool of fatty acids available for assembly into lipoprotein triglycerides by intestinal cells.10 However, as far as we are aware, there are no studies showing the effect of insulin resistance on de novo lipogenesis in the human intestine.
With these precedents, the aims of this study were, first, to determine the relation between insulin resistance/T2DM and intestinal de novo lipogenesis and lipoprotein synthesis in morbidly obese subjects, and second, to study the effect of insulin/glucose on intestinal de novo lipogenesis and lipoprotein synthesis.
Materials and methods
Subjects
The study was undertaken in 45 morbidly obese subjects who underwent a Roux-en-Y gastric bypass, and were classified in three groups according to the homeostasis model assessment of insulin resistance (HOMA-IR) level:11, 12, 13 15 subjects with low HOMA-IR (HOMA-IR<4.7) (MO-low-IR), 15 subjects with high HOMA-IR (HOMA-IR>4.7) (MO-high-IR) (both groups without treatment for T2DM), and 15 subjects with T2DM who were only receiving metformin treatment (MO-metf-T2DM). Subjects were excluded if they had T2DM with insulin treatment, had cardiovascular disease, acute inflammatory or infectious disease, or were receiving drugs that could alter the lipid profile or the metabolic parameters at the time of inclusion in the study. All subjects were of Caucasian origin. Samples from subjects were processed and frozen immediately after their reception in the Regional University Hospital Biobank (Andalusian Public Health System Biobank). All the participants gave their written informed consent and the research project was reviewed and approved by the Ethics and Research Committee of Regional University Hospital, Malaga, Spain.
Laboratory Measurements
Blood samples were collected after 10–12 h fasting. The serum was separated and immediately frozen at −80 °C until analysis. Serum biochemical variables were measured in duplicate, as previously described.3, 11, 12, 13 Apo B-48 was analyzed by enzyme immunoassay (ELISA) (Shibayagi Co., Japan). Cross-reaction with human apolipoprotein B-100 was less than the lower detection limit. The intra- and inter-assay CV was 3.5% and 5.7%, respectively. The sensitivity of the technique was 2.5 ng/ml. Apo A-IV was analyzed by ELISA (Millipore Corporation, Billerica, MA, USA). The intra- and inter-assay CV was 4.6% and 12.2%, respectively. The sensitivity of the technique was 0.078 μg/ml. The HOMA-IR was calculated with the following equation: HOMA-IR=fasting insulin (μIU/ml) × fasting glucose (mmol/l)/22.5.
Jejunal Biopsy Samples
The jejunal biopsy samples were obtained from morbidly obese subjects (n=45) during the bariatric surgery, 40 cm from the ligament of Treitz.3 The mucosa was washed with physiological saline solution, scraped, immediately frozen in liquid nitrogen, and maintained at −80 °C until analysis. Other jejunal biopsies from the morbidly obese subjects group with low HOMA-IR were taken to isolate intestinal epithelial cells (IEC) (n=6).
IEC Isolation and Incubation
All reagents were from Sigma-Aldrich (Sigma-Aldrich, St Louis, MO, USA) unless otherwise specified. IEC were isolated as previously described.14 In brief, jejunal biopsy samples were obtained from morbidly obese subjects with low HOMA-IR (n=6) during the bariatric surgery and washed in a solution composed of 115 mM NaCl, 5.4 mM KCl, 0.96 mM NaH2PO4, 26.19 mM NaHCO3, 5.4 mM glucose, and 1% penicillin/streptomycin adjusted to pH 7.4. Then, the biopsy samples were incubated at 37 °C for 15 min in a solution composed of 67.5 mM NaCl, 1.5 mM KCl, 0.96 mM NaH2PO4, 26.19 mM NaHCO3, 27 mM C6H5Na3O7·2H2O, 5.4 mM glucose, and 1% penicillin/streptomycin adjusted to pH 7.4. After the incubation the solution was discarded and the biopsy samples were incubated at 37 °C for 15 min in a solution composed of 67.5 mM NaCl, 1.5 mM KCl, 0.96 mM NaH2PO4, 26.19 mM NaHCO3, 27 mM C6H5Na3O7·2H2O, 3 mM EDTA, 2 mM dithiothreitol, 5.4 mM glucose, and 1% penicillin/streptomycin adjusted to pH 7.4. After the incubation, the samples were well mixed, the biopsy samples were discarded and the supernatant was centrifuged at 400 g for 2 min. The supernatant was discarded after centrifuging and the pellet was re-suspended in calcium-free Dulbecco’s modified Eagle’s medium (DMEM).
IEC were washed twice with DMEM and cultured in 24-well plates (200,000 cells/well) with DMEM supplemented with 1% fetal bovine serum, 1% l-glutamine, 1% penicillin and streptomycin at 37 °C, and 5% CO2 for 3 h. Tests were performed in different conditions: 5.5 mM glucose, 5.5 mM glucose+100 nM insulin, 25 mM glucose, and 25 mM glucose+100 nM insulin. Each treatment was performed in triplicate. Following these treatments, cell suspensions were centrifuged at 800 g for 3 min, the supernatant was discarded and the cell pellets immediately frozen at −80 °C until analysis.
IEC Characterization by Flow Cytometry
IEC were isolated as described above (n=3). Subsequently, cells were washed twice with PBS and the number and frequency of IEC was determined by flow cytometry. Cells were kept on ice during 30 min and then were incubated with a rat monoclonal IgG antihuman A33-Alexa Fluor 488 (Clone # 402104) (R&D Systems, Minneapolis, MN),15 and a monoclonal IgG antihuman-CD45-APC, a marker of all leukocyte groups (BD Pharmingen, San Jose, CA). Appropriate isotype controls were included (IgG2a-Alexa Fluor 488 (R&D Systems) and IgG1-APC, (BD Pharmingen)) as described16 and analyzed by flow cytometry in a FACS CANTO II (Becton-Dikinson Biosciences, San Jose, CA, USA). Data were analyzed using FlowJo Software. Results were expressed as the percentage of positive cells and the mean fluorescence intensity.
IEC Viability
A trypan blue exclusion test was assessed to determine the number of viable cells.17 Also, a cell viability assay was performed in triplicate using the CellTiter-Glo Luminescent Cell Viability Assay (Promega, Southampton, UK) according to the manufacturer’s instructions.
RNA Extraction and RT-PCR
Total RNA isolation from jejunal biopsy samples and IEC incubations were performed using RNeasy Mini Kit (QIAGEN Science, Hilden, Germany), according to the manufacturer’s instructions. The purity and the integrity of the RNA were determined as described.18 cDNA was synthesized by retrotranscription using the M-MLV retrotranscriptase (Sigma-Aldrich). Gene expression levels were analyzed in triplicate by quantitative real-time reverse transcriptase-PCR using ABI 7500 Fast Real-Time PCR System (Applied Biosystems, Foster City, CA, USA).19 Primers for the PCR reaction were designed based on NCBI database sequences and obtained from Proligo (Sigma-Aldrich): 18s: NM_014473.2; apo A-IV: NM_000482.3; microsomal triglyceride transfer protein (MTTP): NM_000253; diacylglycerol acyltransferase 2 (DGAT2): NM_012079.4; ACLY: NM_001096.2; pyruvate dehydrogenase (lipoamide) beta (PDHB): NM_000925.3; ACC1: NM_133439.2; SREBP-1c: NM_001005291 and FAS: NM_004104.4. Relative expression was calculated using the comparative Ct method (2−ΔΔCt)20 and expression results were given as the expression ratio relative to 18S gene expression.
Statistical Analysis
The statistical analysis was done with SPSS (Version 11.5 for Windows; SPSS, Chicago, IL). Differences between two groups were made with the Mann–Whitney test. Differences between different conditions in the IEC incubations were made with a repeated-measure ANOVA. Partial correlation coefficients (adjusted by body mass index (BMI)) were calculated to estimate the associations between variables. Multiple linear regressions were also used to determine the associations between variables. Values were considered to be statistically significant when the P≤0.05. The results were given as the mean±s.d., or mean±s.e.m. in figures.
Results
Anthropometric and Biochemical Characteristics
The biochemical and anthropometric characteristics of the subjects included in this study are shown in Table 1.
Jejunal mRNA Expression Levels
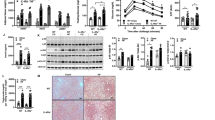
In the MO-high-IR group, apo A-IV (P=0.016), ACLY (P=0.045), PDHB (P=0.043), and SREBP-1c (P=0.019) mRNA expression levels were significantly higher, and ACC1 (P=0.011) and FAS (P=0.042) significantly lower than in the MO-low-IR group (Figure 1). When the MO-low-IR and MO-metf-T2DM groups were compared, these significant differences were maintained in ACLY (P=0.043) and PDHB (P=0.042) mRNA expression levels (Figure 1). Between the MO-high-IR and MO-metf-T2DM groups, there are only significant differences in FAS mRNA expression (P=0.049) (Figure 1).
mRNA expression levels of genes studied in jejunum of morbidly obese subjects with low (MO-low-IR) and high insulin resistance (MO-high-IR), and in morbidly obese subjects with type 2 diabetes treated with metformin (MO-metf-T2DM): (a) sterol regulatory element-binding protein-1c (SREBP-1c), (b) ATP-citrate lyase (ACLY), (c) pyruvate dehydrogenase (lipoamide) beta (PDHB), (d) acetyl-CoA carboxylase alpha (ACC1), (e) fatty-acid synthase (FAS), (f) apolipoprotein A-IV (apo A-IV), (g) diacylglycerol acyltransferase 2 (DGAT2), and (h) microsomal triglyceride transfer protein (MTTP). The results are given as the mean±s.e.m. Significant differences between MO-low-IR and MO-high-IR groups: *P<0.05. Significant differences between MO-low-IR and MO-metf-T2DM groups: aP<0.05. Significant differences between MO-high-IR and MO-metf-T2DM groups: 1P<0.05.
Association Between Jejunal mRNA Expression and Other Variables
We found different significant associations between anthropometric and biochemical variables and gene mRNA expressions. As the groups differed in their BMI, adjustment for BMI was performed in the correlation analyses. Apo A-IV mRNA expression levels correlated positively with insulin (r=0.500, P=0.013) and HOMA-IR (r=0.358; P=0.046). PDHB mRNA expression levels correlated positively with glucose (r=0.671; P=0.001) and HOMA-IR (r=0.638, P=0.002). ACC1 mRNA expression levels correlated negatively with waist (r=−0.522; P=0.022) and hip circumference (r=−0.503; P=0.028). SREBP-1c mRNA expression levels correlated negatively with waist circumference (r=−0.477; P=0.048) and positively with insulin (r=0.418, P=0.027). ACLY mRNA expression levels correlated positively with glucose (r=0.516, P=0.016) and triglycerides (r=0.508, P=0.037). DGAT2, MTTP, and FAS mRNA expression have not significantly correlated with the anthropometric and biochemical variables (data not shown).
Apo A-IV mRNA expression levels correlated positively with SREBP-1c (r=0.513; P=0.013), ACLY (r=0.564; P=0.023), and FAS (r=0.462; P=0.030) mRNA expression levels. DGAT2 mRNA expression levels correlated positively with SREBP-1c (r=0.480; P=0.013) and FAS (r=0.422, P=0.045) mRNA expression levels. PDHB mRNA expression levels correlated negatively with ACC1 mRNA expression levels (r=−0.554; P=0.009). ACC1 mRNA expression levels correlated positively with FAS mRNA expression levels (r=0.451; P=0.024). Adjustments for BMI were performed in the correlation analyses. There were no significant correlations with the remaining mRNA expression studied (data not shown).
To strengthen the independence of these associations as predictors of mRNA gene expression, multiple regression analysis models were constructed for each gene, being the independent variables those that were significant in the correlations: glucose, insulin, free fatty acids, weight and waist circumference, and SREBP-1c mRNA expression. The mRNA expression of most of the genes studied was associated to insulin and/or glucose levels, and to SREBP-1c mRNA expression (Table 2).
mRNA Expression Levels in Incubated IEC
Given the significant associations found between insulin, glucose, and mRNA gene expression levels, we checked whether insulin could modify the expression of these genes. First, we performed a basal characterization of the cells isolated (Figure 2). IEC were identified as A33+CD45− cells among the living cells obtained from the jejunum of morbidly obese subjects. The mean frequency of A33+CD45− cells represented the 84.2±5.6% of the cellular fraction isolated, expressing a mean fluorescence intensity 35.142. The mean frequency CD45+ cells represented 14.9±3.4%. Before incubations, the number of viable cells determined using the trypan blue exclusion test was 87±5.2%. After 3 h of incubation, the number of viable cells determined using the trypan blue exclusion test was above 80.3±7.2%. After 3 h of incubation, the cell viability measured by the CellTiter-Glo Luminescent Cell Viability Assay was 85.1±4.6% (n=6) respect to before incubations. No significant differences were found in the viability test between before and after incubation. Accordingly, IEC from six morbidly obese subjects with low HOMA-IR were incubated with different concentrations of glucose (5.5 and 25 mM) and insulin (0 and 100 nM).
Phenotypic characterization of cells isolated from jejunum of morbidly obese patients by flow cytometry (n=3). The cells isolated were gated on size (FSC) and singularity (SSC) and IgG antihuman-CD45-APC and IgG antihuman A33-Alexa Fluor 488. Intestinal epithelial cells were identified as A33+CD45− cells. A representative histogram is shown with A33+CD45− cells (filled line) and matched-isotypic control (dashed line). Percentage positivity and mean fluorescence intensity are shown in parentheses.
Regarding the IEC treated with 5.5 mM glucose, the incubation with a high dose of insulin (5.5 mM glucose+100 nM insulin) produced a significant increase of apo A-IV, PDHB, and SREBP-1c mRNA expression, and a decrease of ACLY mRNA expression (Figure 3). The incubation with high levels of glucose (25 mM glucose) produced a significant increase of apo A-IV and SREBP-1c, when compared with IEC treated with 5.5 mM glucose (Figure 3).
mRNA expression levels of genes studied in intestinal epithelial cells (IECs) incubations obtained from jejunum samples of morbidly obese subjects with low insulin resistance (n=6). IEC were isolated and incubated as is described in the Materials and Methods section. Tests were performed in different conditions: 5.5 mM glucose, 5.5 mM glucose+100 nM insulin, 25 mM glucose, and 25 mM glucose+100 nM insulin. The results were expressed as a percentage of 5.5 mM glucose level (100%). The results were given as the mean±s.e.m. aSignificant differences between 5.5 mM glucose and 5.5 mM glucose+100 nM insulin (P<0.05). bSignificant differences between 5.5 mM glucose and 25 mM glucose (P<0.05). cSignificant differences between 5.5 mM glucose and 25 mM glucose+100 nM insulin (P<0.05). dSignificant differences between 5.5 mM glucose+100 nM insulin and 25 mM glucose+100 nM insulin (P<0.05). eSignificant differences between 25 mM glucose and 25 mM glucose+100 nM insulin (P<0.05). (a) sterol regulatory element-binding protein-1c (SREBP-1c), (b) ATP-citrate lyase (ACLY), (c) pyruvate dehydrogenase (lipoamide) beta (PDHB), (d) acetyl-CoA carboxylase alpha (ACC1), (e) fatty-acid synthase (FAS), (f) apolipoprotein A-IV (apo A-IV), (g) diacylglycerol acyltransferase 2 (DGAT2), and (h) microsomal triglyceride transfer protein (MTTP).
In the IEC incubated with high doses of glucose and insulin (25 mM glucose+100 nM insulin), mRNA expression levels of DGAT2, MTTP, apo A-IV, and ACC1 were significantly higher than in the IEC incubated with only a high dose of insulin (5.5 mM glucose+100 nM insulin) (Figure 3). Also, mRNA expression levels of DGAT2, apo A-IV, PDHB, and ACC1 with high doses of glucose and insulin (25 mM glucose+100 nM insulin) were significantly higher than in the IEC incubated with only a high dose of glucose (25 mM glucose), whereas ACLY was significantly lower (Figure 3). In the IEC incubated with high doses of glucose and insulin (25 mM glucose+100 nM insulin), mRNA expression levels of DGAT2, MTTP, apo A-IV, ACC1, PDHB, and SREBP-1c were significantly higher than in the IEC incubated with only 5.5 mM glucose (Figure 3).
Discussion
In this study we found that morbidly obese subjects with higher insulin resistance have an altered mRNA expression of genes involved in de novo lipogenesis and lipoprotein synthesis. This altered mRNA expression is attenuated in those morbidly obese subjects treated with metformin. Also, the expression of most of the genes studied is regulated by glucose and/or insulin levels.
Evidence from previous research indicates that insulin-resistant fructose-fed hamsters develop increased intestinal fatty-acid synthesis.7 In our research we observed that in the jejunum of morbidly obese subjects with high insulin resistance there is a coordinated change in the expression of genes involved in lipid metabolism, e.g., a decreased expression of genes involved in FAS and ACC, an increase in acetyl-CoA synthesis (ACLY and PDHB), and an increased expression of apo A-IV, the gene involved in the synthesis and secretion of chylomicrons.
De novo synthesis of fatty acids in morbidly obese subjects with high insulin resistance seems to be decreased, as ACC1 and FAS mRNA expression is decreased, as it is also shown in other tissues in humans.11, 21 ACC1 catalyzes the carboxylation of acetyl-CoA to malonyl-CoA, the rate-limiting step in fatty-acid synthesis. Thus, decreased malonyl-CoA concentrations as a result of low ACC1 could lead to decreased de novo lipid synthesis.
However, the synthesis of acetyl-CoA, precursor of fatty acids and cholesterol, seems to be increased in morbidly obese subjects with high insulin resistance. Our results showed a significantly higher mRNA expression of ACLY and PDHB in these subjects. These two enzymes are involved in the synthesis of cytosolic and mitochondrial acetyl-CoA, respectively. ACLY catalyzes the generation of cytosolic acetyl-CoA from citrate and it is the preferable pathway to synthesize acetyl-CoA in mammalian cells. This increase of ACLY mRNA expression could derive from an excess of citrate found in situations with high free fatty acids and glucose levels,22 as in the morbidly obese subjects with high insulin resistance.23 This increase in citrate has also been associated with a decrease in the concentration of malonyl-CoA,22 as our results also suggest (lower ACC1 mRNA expression).
In our research, we found neither a significant increase of the expression of genes involved in the triglyceride synthesis (DGAT2)24 nor transfer of lipid to lipoproteins (MTTP)25 in morbidly obese subjects with high insulin resistance or in diabetic condition, when they are compared with those with low insulin resistance. In obese subjects, a previous study has shown that DGAT activity was not different according to the diabetic status in visceral or subcutaneous adipose tissue.26 At the same time, the results of the expression of intestinal MTTP are contradictory. A previous study conducted in humans showed no significant changes in intestinal MTTP expression in morbidly obese subjects with T2DM,3 whereas other studies show that there is an increase in intestinal MTTP expression in a rabbit model of diabetes27 and in insulin-resistant non-diabetic Zucker obese fa/fa rats.28 However, there is little information regarding MTTP in humans. On the other hand, apo A-IV mRNA expression levels were significantly higher in those subjects with high insulin resistance. Apo A-IV is a lipid-binding protein that is incorporated into the nascent chylomicrons and enhances basolateral triglyceride secretion in a dose-dependent manner.6 This increase found in apo A-IV mRNA expression is according to other research showing an increase in the assembly and secretion of apoB48-containing lipoproteins in insulin-resistant states in humans with insulin resistance29 but with a lower content in triglycerides. Although the expression and synthesis of apo A-IV is very specific for intestine, no significant correlation was found between apo A-IV mRNA expression and serum levels. We do not know the reason of this discrepancy, but the clearance of chylomicrons and high-density lipoproteins (HDL) can affect serum apo A-IV concentration. Apo A-IV is found in plasma on chylomicrons and HDL, but 40–50% circulates lipid free.30 Its levels could be modified in other pathologies not analyzed in this study, such as coronary artery disease and renal impairment.31, 32
SREBP-1c is a transcription factor involved in the regulation of lipogenesis.9 Our results show that SREBP-1c mRNA expression is increased in the jejunum of morbidly obese subjects with a high insulin resistance, as is also shown in insulin-resistant animals.10 This increased expression may have resulted from the increased insulinemia,33 as our results also suggest. Using different multiple regression models, we also found that both, SREBP-1c and/or insulin might be involved in the explanation of mRNA levels of ACC1, MTTP, DGAT2, PDHB, apo A-IV, and FAS. It is known that SREBP-1c can regulate ACC1, FAS, and MTTP,9, 34, 35 and insulin can regulate SREBP-1c,33 FAS,36 apo A-IV,37 and DGAT2.38 Therefore, insulin may be involved in the regulation of these genes, directly or through SREBP-1c. However, other transcription factors can also be involved in the intestinal regulation of levels of these enzymes.
An important finding is that metformin slightly modified the mRNA expression of apo A-IV, ACC1, and SREBP-1c. Metformin is a widely used drug for the treatment of T2DM. Metformin treatment improves the expression of these genes to the point of the disappearing of the significant differences with respect to morbidly obese subjects with low insulin resistance. To our knowledge, there is no research on the effect of metformin on intestinal SREBP-1c and apo A-IV. With respect to SREBP-1c, and according to our results, metformin treatment has been found to downregulate SREBP-1c in L6 muscle cells.39 However, our results are not according to research made in 3T3-L1 adipocytes, in which metformin reduces ACC1 protein.40 These differences could be due to the fact that the regulation of the metabolism of different tissues may differ. Together, our results show a slight beneficial effect of metformin on the restitution of intestinal lipid/lipoprotein homeostasis.
We have also found that the expression of the genes studied is regulated by glucose and/or insulin in the IEC of morbidly obese subjects with low insulin resistance. We have found a higher mRNA expression of DGAT2, MTTP, and apo A-IV in the jejunum from morbidly obese subjects with low insulin resistance when glucose and insulin are at a high concentration, like in postprandial condition, suggesting that there could be a higher synthesis of triglyceride-rich lipoproteins. It is known that apo A-IV synthesis and secretion by the gastrointestinal tract are upregulated by insulin,37 and DGAT2 mRNA expression is specifically increased by insulin.38 Also, an increase in de novo FAS could be found because of an increase of ACC1 mRNA expression, although FAS mRNA expression is not increased. ACC1 gene has an inducible expression in response to insulin.41 However, in a situation with only high levels of glucose or insulin, there is only a higher apo A-IV mRNA expression, suggesting a higher synthesis of triglyceride-rich lipoproteins, but without an increase of its triglyceride content, owing to the absence of an increase of DGAT2, MTTP, and ACC1. On the other hand, PDHB, SREBP-1c, and ACLY behave in an opposite manner in response to insulin. Although PDHB and SREBP-1c mRNA expression is increased when there is a high insulin concentration, ACLY expression is decreased. PDHB and SREBP-1c are two enzymes upregulated by insulin.33, 42 SREBP-1c is involved in the lipogenic effect of chronic hyperinsulinemia,9 and the activation of PDH complex favors glucose oxidation and improves blood glucose control.43 Regarding ACLY, insulin decreased the ACLY mRNA expression, as it has been shown in previous research in which ACLY was downregulated in an insulin dose-dependent manner.44
Several limitations to this research must be acknowledged. First, a large number of proteins and enzymes can contribute to de novo lipogenesis and to the synthesis of triglyceride-rich lipoproteins. In this research we measured only the mRNA expression of several enzymes, but we think that these enzymes may be the most interesting. Another limitation of this research is that we did not have sufficient samples to analyze protein levels or enzymatic activity. Also, we have found a discrepancy between the gene expression in the in vitro experiments and the in vivo study. This could be because of the fact that in the in vitro experiment, the subjects have a low insulin resistance, or it could be due to other factors not studied. On the other hand, it would have been interesting to introduce a control group of lean patients for the tissue analysis, and cells from lean patients for the in vitro approach, which would have responded better to glucose and insulin compared with morbidly obese patients. However, we have not been able to obtain jejunal biopsies from the same location of healthy lean patients.
In conclusion, the jejunum of morbidly obese subjects with high insulin resistance has a decreased expression of genes involved in de novo synthesis of fatty acids and an increased expression of genes involved in the acetyl-CoA and lipoprotein synthesis. Lipogenesis is a synchronized mechanism, and the alteration of the intestinal environment, like in morbidly obese subjects with high insulin resistance, can cause alterations in the expression of different enzymes. Also, the results of this research show a slight beneficial effect of metformin on the restitution of intestinal lipid/lipoprotein homeostasis. Moreover, we show that the effects that high levels of insulin and/or glucose have on the IEC of morbidly obese subjects with low insulin resistance are different with respect to the mRNA expression pattern of the jejunum samples isolated from morbidly obese subjects with high insulin resistance.
References
Haffner SM, Foster DM, Kushwaha RS et al. Retarded chylomicron apolipoprotein-B catabolism in type 2 (non-insulin-dependent) diabetic subjects with lipaemia. Diabetologia 1984;26:349–354.
Castillo M, Hortal JH, García-Fuentes E et al. Coconut oil affects lipoprotein composition and structure of neonatal chicks. J Biochem 1996;119:610–616.
Soriguer F, García-Serrano S, Garrido-Sánchez L et al. Jejunal wall triglyceride concentration of morbidly obese persons is lower in those with type 2 diabetes mellitus. J Lipid Res 2010;51:3516–3523.
Martins IJ, Sainsbury AJ, Mamo JC et al. Lipid and apolipoprotein B48 transport in mesenteric lymph and the effect of hyperphagia on the clearance of chylomicron-like emulsions in insulin-deficient rats. Diabetologia 1994;37:238–246.
Otokozawa S, Ai M, Diffenderfer MR et al. Fasting and postprandial apolipoprotein B-48 levels in healthy, obese, and hyperlipidemic subjects. Metabolism 2009;58:1536–1542.
Lu S, Yao Y, Cheng X et al. Overexpression of apolipoprotein A-IV enhances lipid secretion in IPEC-1 cells by increasing chylomicron size. J Biol Chem 2006;281:3473–3483.
Haidari M, Leung N, Mahbub F et al. Fasting and postprandial overproduction of intestinally derived lipoproteins in an animal model of insulin resistance. Evidence that chronic fructose feeding in the hamster is accompanied by enhanced intestinal de novo lipogenesis and ApoB48-containing lipoprotein overproduction. J Biol Chem 2002;277:31646–31655.
Duez H, Lamarche B, Uffelman KD et al. Hyperinsulinemia is associated with increased production rate of intestinal apolipoprotein B-48-containing lipoproteins in humans. Arterioscler Thromb Vasc Biol 2006;26:1357–1363.
Horton JD, Goldstein JL, Brown MS . SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest 2002;109:1125–1131.
Federico LM, Naples M, Taylor D et al. Intestinal insulin resistance and aberrant production of apolipoprotein B48 lipoproteins in an animal model of insulin resistance and metabolic dyslipidemia: evidence for activation of protein tyrosine phosphatase-1B, extracellular signal-related kinase, and sterol regulatory element-binding protein-1c in the fructose-fed hamster intestine. Diabetes 2006;55:1316–1326.
Tinahones FJ, Garrido-Sanchez L, Miranda M et al. Obesity and insulin resistance-related changes in the expression of lipogenic and lipolytic genes in morbidly obese subjects. Obes Surg 2010;20:1559–1567.
Garcia-Fuentes E, Murri M, Garrido-Sanchez L et al. PPARgamma expression after a high-fat meal is associated with plasma superoxide dismutase activity in morbidly obese persons. Obesity (Silver Spring) 2010;18:952–958.
García-Serrano S, Moreno-Santos I, Garrido-Sánchez L et al. Stearoyl-CoA desaturase-1 is associated with insulin resistance in morbidly obese subjects. Mol Med 2011;17:273–280.
Cartwright IJ, Higgins JA . Isolated rabbit enterocytes as a model cell system for investigations of chylomicron assembly and secretion. J Lipid Res 1999;40:1357–1365.
Johnstone CN, Tebbutt NC, Abud HE et al. Characterization of mouse A33 antigen, a definitive marker for basolateral surfaces of intestinal epithelial cells. Am J Physiol Gastrointest Liver Physiol 2000;279:G500–G510.
Rodriguez-Pena R, Lopez S, Mayorga C et al. Potential involvement of dendritic cells in delayed-type hypersensitivity reactions to beta-lactams. J Allergy Clin Immunol 2006;118:949–956.
Rodriguez-Pacheco F, Garcia-Serrano S, Garcia-Escobar E et al. Effects of obesity/fatty acids on the expression of GPR120. Mol Nutr Food Res 2014;58:1852–1860.
Garrido-Sánchez L, Vendrell J, Fernández-García D et al. De novo lipogenesis in adipose tissue is associated with course of morbid obesity after bariatric surgery. PLoS One 2012;7:e31280.
Gutiérrez-Repiso C, Velasco I, Garcia-Escobar E et al. Does dietary iodine regulate oxidative stress and adiponectin levels in human breast milk? Antioxid Redox Signal 2014;20:847–853.
Garrido-Sánchez L, García-Fuentes E, Fernández-García D et al. Zinc-alpha 2-glycoprotein gene expression in adipose tissue is related with insulin resistance and lipolytic genes in morbidly obese patients. PLoS One 2012;7:e33264.
Ortega FJ, Mayas D, Moreno-Navarrete JM et al. The gene expression of the main lipogenic enzymes is downregulated in visceral adipose tissue of obese subjects. Obesity (Silver Spring) 2010;18:13–20.
Ruderman NB, Saha AK, Vavvas D et al. Malonyl-CoA, fuel sensing, and insulin resistance. Am J Physiol 1999;276:E1–E18.
Soriguer F, García-Serrano S, García-Almeida JM et al. Changes in the serum composition of free-fatty acids during an intravenous glucose tolerance test. Obesity (Silver Spring) 2009;17:10–15.
Yu YH, Ginsberg HN . The role of acyl-CoA:diacylglycerol acyltransferase (DGAT) in energy metabolism. Ann Med 2004;36:252–261.
Hussain MM, Shi J, Dreizen P . Microsomal triglyceride transfer protein and its role in apoB-lipoprotein assembly. J Lipid Res 2003;44:22–32.
Hou XG, Moser S, Sarr MG et al. Visceral and subcutaneous adipose tissue diacylglycerol acyltransferase activity in humans. Obesity (Silver Spring) 2009;17:1129–1134.
Phillips C, Bennett A, Anderton K et al. Intestinal rather than hepatic microsomal triglyceride transfer protein as a cause of postprandial dyslipidemia in diabetes. Metabolism 2002;51:847–852.
Phillips C, Owens D, Collins P et al. Microsomal triglyceride transfer protein: does insulin resistance play a role in the regulation of chylomicron assembly? Atherosclerosis 2002;160:355–360.
Duez H, Lamarche B, Valéro R et al. Both intestinal and hepatic lipoprotein production are stimulated by an acute elevation of plasma free fatty acids in humans. Circulation 2008;117:2369–2376.
Wang F, Kohan AB, Lo CM et al. Apolipoprotein A-IV: a protein intimately involved in metabolism. J Lipid Res 2015;56:1403–1418.
Ezeh B, Haiman M, Alber HF et al. Plasma distribution of apoA-IV in patients with coronary artery disease and healthy controls. J Lipid Res 2003;44:1523–1529.
Lingenhel A, Lhotta K, Neyer U et al. Role of the kidney in the metabolism of apolipoprotein A-IV: influence of the type of proteinuria. J Lipid Res 2006;47:2071–2079.
Hegarty B, Bobard A, Hainault I et al. Distinct roles of insulin and liver X receptor in the induction and cleavage of sterol regulatory element-binding protein-1c. Proc Natl Acad Sci USA 2005;102:791–796.
Zhao LF, Iwasaki Y, Zhe W et al. Hormonal regulation of acetyl-CoA carboxylase isoenzyme gene transcription. Endocr J 2010;57:317–324.
Sato R, Miyamoto W, Inoue J et al. Sterol regulatory element-binding protein negatively regulates microsomal triglyceride transfer protein gene transcription. J Biol Chem 1999;274:24714–24720.
Griffin MJ, Sul HS . Insulin regulation of fatty acid synthase gene transcription: roles of USF and SREBP-1c. IUBMB Life 2004;56:595–600.
Hanniman EA, Lambert G, Inoue Y et al. Apolipoprotein A-IV is regulated by nutritional and metabolic stress: involvement of glucocorticoids, HNF-4a, and PGC-1a. J Lipid Res 2006;47:2503–2514.
Meegalla RL, Billheimer JT, Cheng D . Concerted elevation of acyl-coenzyme A:diacylglycerol acyltransferase (DGAT) activity through independent stimulation of mRNA expression of DGAT1 and DGAT2 by carbohydrate and insulin. Biochem Biophys Res Commun 2002;298:317–323.
Bi Y, Wu W, Shi J et al. Role for sterol regulatory element binding protein-1c activation in mediating skeletal muscle insulin resistance via repression of rat insulin receptor substrate-1 transcription. Diabetologia 2014;57:592–602.
Boyle JG, Logan PJ, Jones GC et al. AMP-activated protein kinase is activated in adipose tissue of individuals with type 2 diabetes treated with metformin: a randomised glycaemia-controlled crossover study. Diabetologia 2011;54:1799–1809.
Brownsey RW, Boone AN, Elliott JE et al. Regulation of acetyl-CoA carboxylase. Biochem Soc Trans 2006;34:223–227.
Wieland OH . The mammalian pyruvate dehydrogenase complex: structure and regulation. Rev Physiol Biochem Pharmacol 1983;96:123–170.
Mayers RM, Butlin RJ, Kilgour E et al. AZD7545, a novel inhibitor of pyruvate dehydrogenase kinase 2 (PDHK2), activates pyruvate dehydrogenase in vivo and improves blood glucose control in obese (fa/fa) Zucker rats. Biochem Soc Trans 2003;31:1165–1167.
Polakof S, Médale F, Larroquet L et al. Insulin stimulates lipogenesis and attenuates Beta-oxidation in white adipose tissue of fed rainbow trout. Lipids 2011;46:189–199.
Acknowledgements
CIBER Fisiopatología de la Obesidad y Nutrición (CIBEROBN) and CIBER de Diabetes y Enfermedades Metabólicas Asociadas (CIBERDEM) are ISCIII projects. This work was supported in part by a grant from the Instituto de Salud Carlos III, Spain (PI12/00338). This study has been co-funded by FEDER funds.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Gutierrez-Repiso, C., Rodriguez-Pacheco, F., Garcia-Arnes, J. et al. The expression of genes involved in jejunal lipogenesis and lipoprotein synthesis is altered in morbidly obese subjects with insulin resistance. Lab Invest 95, 1409–1417 (2015). https://doi.org/10.1038/labinvest.2015.115
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/labinvest.2015.115
This article is cited by
-
Intestinal lipid absorption and transport in type 2 diabetes
Diabetologia (2022)
-
The vital role of ATP citrate lyase in chronic diseases
Journal of Molecular Medicine (2020)
-
Actions of metformin and statins on lipid and glucose metabolism and possible benefit of combination therapy
Cardiovascular Diabetology (2018)