Abstract
Esophagus squamous cell carcinoma (ESCC) is one of the most deadly malignances because of its high frequency of metastasis. Given the associations of MUC1 with ESCC and tumor metastasis, we explored a potential role of MUC1 in ESCC metastasis. Among 40 ESCC and 20 paired normal tissue specimens examined, we found a significant increase of MUC1 expression in ESCC and more importantly, that expression of MUC1 and MMP13 are strongly correlated in patients who had lymph node metastasis. Studies with cell models indicated that overexpression of MUC1 upregulates the expression of MMP13, leading to increased cell migration. In support of a mode of transcriptional regulation, promoter analysis revealed that MUC1 stimulates MMP13 expression through the Runx-2-binding site. The link of MUC1 to cell motility was further confirmed by the finding that depletion of MUC1 resulted in reduced expression of MMP13 and cell migration, invasion and adhesion. Moreover, the loss of cell metastatic potential was rescued by overexpression of MMP13 completely. Collectively, our findings indicate that MUC1 contributes to ESCC metastasis by stimulating MMP13 expression, suggesting MUC1 as a novel diagnostic biomarker and therapeutic target in ESCC.
Similar content being viewed by others
Main
Esophageal squamous cell carcinoma (ESCC) frequently exhibits extensive local invasion or regional lymph node metastasis at the time of initial diagnosis; therefore, it is one of the most common aggressive diseases with poor outcome.1 Tumor invasion and metastasis involve degradation of different components of the extracellular matrix and require the actions of proteolytic enzymes, such as matrix metalloproteinases (MMPs), which are produced either by the tumor cells or surrounding stromal cells.2, 3 MMP13 is a highly regulated zinc-dependent endopeptidase and has been reported to be associated with vascular invasion and lymph node metastasis in ESCC.4 Mechanisms involved in regulation of MMP13 in ESCC are likely complex and poorly understood.
Mucins are high-molecular-weight glycoproteins that have been identified as markers of adverse prognosis and as attractive therapeutic targets.5 MUC1, one of transmembrane mucins, is normally expressed in esophageal epithelium. Patients with MUC1 high expression often appear with advanced stage or lymph node metastasis suggesting correlation of the MUC1 expression and the invasion or metastasis of ESCC.6 In this study, we investigated the expression of MUC1 and MMP13 in ESCC patients and the potential functional relationship in tumor metastasis and prognosis.
MATERIALS AND METHODS
Tissue Sample Collection
A total of 40 paraffin-embedded archival specimens of primary ESCC cases were enrolled in this study: 20 with lymph node metastasis and 20 without lymph node metastasis. A total of 20 paired normal esophageal tissue specimens distant from the cancerous lesion in patients without lymph node metastasis were used as control. These patients did not receive any preoperative adjuvant radiation or chemotherapy.
Immunohistochemistry Staining and Quantitative Analysis
The paraffin-embedded sections were stained with primary anti-MUC1 N-ter (DF3),7 or anti-MMP13 (Abcam, Cambridge, UK) antibody and horseradish peroxidase-labeled immunoglobulin (Jingmei Biotech Co., Ltd, Shenzhen, China). Images were obtained at × 200 magnification.
Stained slides were scored by two blinded, independent observers. Integrated optical density was measured in three random fields from each patient using Image-Pro Plus 6.0. Values were represented as mean±s.d.
Cell Culture
Rat 3Y1 fibroblasts8 were grown in DMEM with 1.6 μg/ml puromycin, 10% heat-inactivated fetal bovine serum (FBS), 100 U/ml penicillin, 100 μg/ml streptomycin and 2 mmol/l L-glutamine. Human Ec1.71 esophageal squamous carcinoma cells9 were grown in DMEM with 10% heat-inactivated FBS, 100 U/ml penicillin, 100 μg/ml streptomycin and 2 mmol/l L-glutamine.
Plasmid Construction
MMP13-Luciferase reporters were prepared based on plasmid pGL3-Basic (Promega, Madison, WI, USA). The promoter fragments were generated by PCR amplification of human genomic DNA with the following forward primers:10 MMP13 (–1004) 5′-AAGGTACCCTGCAGCCCTAGTTTTCTTGG-3′, MMP13 (–687) 5′-AAGGTACCTCCCTGTTTGTGCTAGCC-3′, MMP13 (–402) 5′-GGGGTACCTCTAGAATCAGTACTAAGTTT-3′, MMP13 (–56) 5′-AAGGTACCAAGTGATGACTCACCATTG-3′ and reverse primer: MMP13R 5′-TTAGATCTTTGAATGGTGATGCCTGG-3′. The PCR products were then cloned into pGL3-Basic using KpnI and BglII sites. Mutations within the Runx-2 and Ets-binding sites were made using QuikChange Site-Directed Mutagenesis Kit (Strategene, Cedar Creek, TX, USA) to mutate 5′-AACCACA-3′ to 5′-ACTAACA-3′ for Runx-2-site, 5′-AGGAAG-3′ to 5′-AAAAAG-3′ for Ets-site. The changed sequences are highlighted in bold.
The full-length ORF (1416 bp) of human MMP13 was amplified by PCR from the cDNA of an esophageal tumor tissue using forward primer: 5′-AATAGATCTCATCCAGGGGTCCTGGCT-3′; and reverse primer: 5′-GCCGTCGACTTAACACCACAAAATGGAA-3′. The PCR product was then cloned into pBabe retroviral vector using BglII and salI sites (Addgene, Cambridge, MA, USA). The constructs were confirmed by DNA sequencing and restriction enzyme digestion.
Transfection and Luciferase Assay
Cells were transiently transfected with pGL3-MMP13-Luciferase reporters and pGL3-TK-Renilla using FuGene HD (Roche Diagnostics, Indianapolis, IN, USA). Luciferase assays were performed with the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA) at 36 h after transfection. The transfection efficiency was normalized with Renilla luciferase activity.
Immunoblot Analysis
Patient tissues or subconfluent cells were collected and total cell lysates were prepared in lysis buffer.8 Proteins were separated by SDS-PAGE, transferred to nitrocellulose membranes and probed with anti-MUC1-N (DF3),7 anti–MUC1-C (Ab-5; NeoMarkers, Fremont, CA, USA), anti-MMP13, anti-Vinculin (Abcam, Cambridge, UK) and anti-β-actin (AC-15; Sigma, St Louis, MO, USA).
Real-Time PCR
Quantitative PCR was carried out with SYBR Green PCR kit according to the manufacturer's instructions (ToYoBo Co., LTD, Osaka, Japan). Amplifications were performed in ABI PRISM 7500 Sequence Detection System (Applied Biosystems). Relative transcript quantities were calculated using the ΔΔCt method with GAPDH as the endogenous reference gene. Each sample was repeated three times.
Zymography Assay
Zymography assay was performed as described.11 In brief, concentrated conditioned media were mixed with non-reducing sample buffer, and separated in SDS-PAGE containing 1 mg/ml gelatin without heating. The gel was subjected to wash with 2.5% Triton X-100 to remove SDS, rinse with 50 mM Tris-HCl, pH 7.6, and then incubated with developing buffer (50 mM Tris-HCl pH 7.6, 5 mM CaCl2, 200 mM NaCl) for 20 h at 37 °C. The zymographic activities were revealed by staining with 1% Coomassie Blue. The sample was also loaded into SDS-PAGE and stained with 1% Coomassie Blue as loading control.
Wound-Healing Assay
Cells were grown in DMEM with 10% FBS in six-well plates until 95% confluence. A straight ‘wound’ was made by scratching with a plastic pipette tip. After culture in FBS-free medium for 46 h, cell migration was analyzed using light microscopy. The distance of the scratch at the same position was measured by using the Image-Pro Plus software. The migration rate was calculated by the following formula: % Migration = (the length of initial wound – the length of wound after 46 h) × 100/the length of initial wound. All experiments were repeated three times independently.
Transwell Migration Assay
Cells (1 × 104 ) were seeded in 8-μm porous transwell chambers (Corning Incorporated, Corning, NY, USA) in DMEM without serum and incubated in 24-well-plates with 10% FBS supplemented DMEM for 16 h. Transmigration cells were stained with 0.1% crystal violet and evaluated using microscopy.
Silencing of MUC1
Two siRNA sequences targeting MUC1 (siRNA-A, 5′-AAGGTACCATCAATGTCCACG-3′, or siRNA-B, 5′-AAGTTCAGTGCCCAGCTCTAC-3′) and a control sequence (5′-CGCTTACCGATTCAGAATGG-3′)12 were cloned into siRNA expression vector pRNA6.1 using BamHI and HindIII sites. Ec1.71 cells were transfected with the siRNA plasmids in the presence of Lipofectamine. Stable transfectants were selected with 1.5 mg/ml G418.
Cell Infection
To obtain stable MMP13-expressing cell line, 293T-packaging cells plated in 100-mm dishes were co-transfected with the pBabe-Flag-MMP13 or empty retroviral vector and the packaging plasmid using Fugene HD transfection agent (Roche). Ec1.71 MUC1-siRNA-B stable cell line was infected with the retroviral supernatant twice in the presence of polybrene. Cells were selected with 0.25 μg/ml puromycin for 1 week after 48 h infection.
Invasion Assay
The upper chamber of each transwell was coated with Matrigel (BD Biosciences, MA, USA) 1:6 diluted with DMEM at 37 °C for 3 h. Cells (2 × 104) were seeded in upper chambers in DMEM and incubated in 24-well-plates with 10% FBS supplemented DMEM. After 36 h of incubation, cells remaining on the upper surface of the membrane were removed with a cotton swab. Cells that invaded through the Matrigel-precoated membrane filter were fixed, stained and counted using a microscope.
Cell Adhesion Assay
In all, 1 × 103 single-cell were seeded into 96-well plates precoated with Matrigel 50 mg/l (BD Biosciences). After 1 h incubation at 37 °C, the wells were washed three times with PBS to remove non-adherent cells. The adherent cells were fixed by methanol for 15 min, and then stained with 0.1% crystal violet for 30 min at 25 °C. Plates were washed three times in distilled water and dried. Stained cells were counted under the phase contrast microscope.
Aggregation Assay
The cell aggregation assay was performed essentially as described.13 Briefly, a single-cell suspension at about 1 × 105 per 0.5 ml in serum-free DMEM was placed in polystyrene microtubes rotated at 80 r.p.m. for 1 h at 37 °C . The aggregation process was stopped using 2% (v/v) glutaraldehyde. The percentage of aggregated cells was calculated as (N0-Nt)/N0 × 100%, where Nt is the number of single cells after incubation 1 h at 37 °C and N0 is the total number of the cells added to the Medium.
Statistical analysis
All analyses were conducted using SAS 9.12 (SAS Institute, Inc., Cary, NC, USA). Analysis of survival was performed using Kaplan–Meier method and compared using log-rank test. Fisher exact test and Student's t-test were used for between-group comparisons. A probability value of <0.05 was considered to be of significance.
RESULTS
Increased Expression of MUC1 and MMP13 is Associated with Lymph Node Metastasis in ESCC
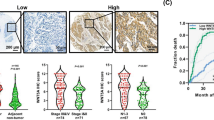
Results of immunohistochemistry (IHC) staining showed that MUC1 was weakly expressed in normal esophageal epithelium, whereas little MMP13 was detectable (Figure 1a left panel). On the contrary, MUC1 and MMP13 were moderately or strongly expressed in most ESCC specimens. Significantly, MUC1 and MMP13 were found to be coexpressed in 16 tumor tissues with lymph node metastasis (Figure 1a middle panel, Figure 1b and Table 1). Interestingly, MUC1 was less or non-detectable in three of the other four lymph node positive samples that expressed little MMP13 (Figure 1a right panel). Both MUC1 and MMP13 had a significantly higher incidence of overexpression in ESCC with lymph node metastasis than those without lymph node metastasis (P<0.05 for MUC1, P<0.01 for MMP13, P<0.005 for co-overexpresssion of MUC1 and MMP13; Figure 1b and Table 1). Among all the cases examined, patients with lymph node metastasis showed significantly poorer survival than patients without lymph node metastasis (P<0.01, Figure 1c). Consistent with these IHC results, western blot analysis on four tumor tissues with lymph node metastasis and four un-matched normal tissues showed that MUC1 protein was present at a considerably higher level in tumor tissues when compared with un-matched normal tissues (Figure 1d). RT-PCR assay on these tissues revealed substantially higher mRNA level of MMP13 in tumor tissues compared with un-matched normal tissues (Figure 1e). These results indicate that ESCC metastasis is associated with significantly increased expression of MUC1 and MMP13, and interestingly, these two proteins seem to coexpress in ESCC.
Increased expression of MUC1 and MMP13 is associated with lymph node metastasis in ESCC. (a) IHC staining of MUC1 and MMP13 on sections from esophageal tumor with lymph node metastasis and normal epithelial tissue from paracancer without lymph node metastasis. Photomicrographs obtained at × 200 magnification. (b) Quantitative analysis of MUC1 and MMP13 expression in all IHC staining. (c) Kaplan–Meier survival curve for patients with or without lymph node metastasis. (d) Lysates from tumor tissues (T1–T4) and un-matched normal tissues (P1–P4) were immunoblotted with antibody against MUC1-N and Vinculin as loading control. (e) Total RNA from tumor tissues (T1–T4) and un-matched normal tissues (P1–P4) were extracted by using Trizol reagent according to the manufacturer's protocol. cDNA was generated by reverse transcription using AMV reverse transcriptase. RT-PCR analysis was performed using primers of MMP13 and GAPDH.
MUC1 Overexpression is Associated with Elevated Levels of MMP13 Protein and mRNA in 3Y1 Cells
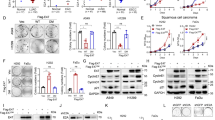
The coexpression of MUC1 and MMP13 in ESCC suggests a potential interaction of these two proteins, which we studied by examining the 3Y1 cells that stably expressed either empty vector or Flag-tagged MUC1-CD containing a 72 amino-acid cytoplasmic domain.8 A 3Y1/Vector and two independent clones of 3Y1/MUC1-CD cells were used. RT-PCR analysis of total RNA showed increased MMP13 mRNA level in 3Y1/MUC1-CD-A and 3Y1/MUC1-CD-B cells when compared with 3Y1/Vector cells (Supplementary Figure S1). Results of real-time PCR also showed that MMP13 mRNA was increased more than threefold in 3Y1/MUC1-CD-A and 3Y1/MUC1-CD-B when compared with 3Y1/Vector (Figure 2a). Consistent with these data, immunoblot analysis of the transfectants showed significantly elevated MMP13 protein in 3Y1/MUC1-CD cells (Figure 2b). Zymography assay indicated higher activity of MMP13 in 3Y1/MUC1-CD cells than 3Y1/Vector cells (Figure 2c). These results demonstrate that MUC1 overexpression is associated with increased MMP13 expression and activity in 3Y1 cell lines.
MUC1-CD co-activates MMP13 transcription. (a) Real-time PCR evaluates mRNA level of MMP13 in 3Y1/Vector (open bar) and 3Y1/MUC1-CD stale cell lines A (solid bar) and B (grey bar). Results are expressed as relative fold of mRNA compared with that obtained in 3Y1/vector cells (assigned value of 1). Columns, mean of three separate experiments; bars, s.d. (b) Lysates from the indicated cells were immunoblotted with antibody against MUC1-C, Mmp13 and β-actin. (c) The culture medium from indicated cells was collected and the activities of Mmp13 were measured by gelatin-zymography (upper). Coomassie blue staining of SDS-PAGE without gelatin as loading control (lower). (d upper) Schematic representation of structure of MMP13 promoter–luciferease reporters. (d lower and e) 3Y1/Vector or 3Y1/MUC1-CD cells were transiently transfected with indicated MMP13 promoter–luciferease reporters. Dual-luciferase reporter assay was performed at 36 h after transfection. Results are expressed as relative fold of luciferease activity normalized with Renilla. Columns represent the mean of three independent transfection experiments; bars, s.d.
MUC1-CD Co-activates MMP13 Transcription
To determine whether MUC1 upregulates MMP13 by activating its transcription, four different fragments of MMP13 promoter were amplified and cloned into pGL3-basic vector (Figure 2d upper). Luciferase activity was analyzed by transient transfection of reporters and pGL3-basic vector control into 3Y1/Vector and 3Y1/MUC1-CD stable cell lines. In comparison with 3Y1/Vector cells, the transcriptional activity of P-402, P-687 and P-1004 reporters were increased in 3Y1/MUC1-CD cells. Importantly, the highest luciferase activity (threefold increase) was detected with the reporter P-402 that contains the Ets site and Runx-2 site (Figure 2d lower). To further map the possible elements on which MUC1 effects, luciferase assay was performed using vectors with either mutated Ets (P-402E) or mutated Runx-2 (P-402R). Results revealed that transcriptional activation of P-402R was substantially decreased in both 3Y1/Vector and 3Y1/MUC1-CD cells when compared with wild-type P-402. By contrast, the activity of P-402E was decreased in 3Y1/Vector, but increased slightly in 3Y1/MUC1-CD (Figure 2e). The results suggest that Runx-2 is the key protein that regulates MMP13 gene expression; MUC1-CD stimulates MMP13 expression through Runx-2. Moreover, we find that with absence of MUC1, Ets modulates MMP13 expression by cooperating with Runx-2. Whereas in the presence of MUC1-CD, it is dispensable to MMP13 expression.
MUC1 Overexpression Enhances Cell Migration
In light of the finding that MUC1 upregulates MMP13 expression, we asked whether MUC1-CD was capable of affecting cell migration. To investigate this, a wound-healing assay was performed with 3Y1/Vector and 3Y1/MUC1-CD cells. Results showed that the closure of the wounds was apparently faster in 3Y1/MUC1-CD cells in which most space along the migrating fronts fused, whereas the wounds remained open in 3Y1/Vector cells (Figure 3a). The migration rate of 3Y1/MUC1-CD cells was found to be significantly higher when compared with that of 3Y1/Vector cells (P<0.001, Figure 3b). The migration capacity of 3Y1 stable transfectants was also assessed in transwell plates. The cell number of transmigration in 3Y1/MUC1-CD was approximately more than twofold of that in 3Y1/Vector control (P<0.001, Figure 3c and d). These results indicate that overexpression of MUC1-CD is associated with enhanced cell migration.
MUC1 overexpression enhances cell migration. (a) 3Y1/Vector or 3Y1/MUC1-CD cells were grown until confluence. ‘Wound’ with a uniform width was made on the monolayer cells by scratching with a plastic pipette tip. The cells were cultured in FBS-free DMEM for another 46 h. Pictures were taken using light microscopy, × 100. (b) Migration rate of 3Y1/Vector (open bar) and 3Y1/MUC1-CD (solid bar) was calculated from three independent experiments as formula: % Migration=(the length of initial wound – the length of wound after 46 h) × 100/the length of initial wound. P<0.001. (c) The indicated cells (1 × 104) were seeded on 8-μm porous transwell chambers. After 16 h of plating, transmigration cells were fixed and stained with crystal violet. Photomicrographs obtained at × 100 magnification. (d) Transmigration cells were counted for each of the indicated cells. Columns, mean number of cells obtained in three independent experiments; bars, s.d.; ***P<0.001.
Depletion of MUC1 Reduces MMP13 Expression and Cell Migration
To determine whether MUC1 enhances cell migration by increasing MMP13 expression, we depleted the expression of MUC1 in an ESCC cell line, Ec1.71 cells by stably transfecting control siRNA (CTL-siRNA), or MUC1-siRNA. Immunoblotting of cell lysates from transfectants exhibited that MUC1 expression was significantly downregulated by MUC1-siRNAs (Figure 4a). In support of MUC1-mediated MMP13 regulation, either MMP13 mRNA or MMP13 activity also decreased in MUC1-siRNAs but not in CTL-siRNA expression cells (Figure 4b and c). In agreement with these data, a transwell migration assay performed with Ec1.71 transfectants showed that the migration cells were markedly decreased by MUC1-siRNAs when compared with CTL-siRNA (Figure 4d). The number of transmigration cells of Ec1.71/MUC1-siRNAs was significantly less than that expressing CTL-siRNA (P<0.001, Figure 4e). Consistent with the finding in Ec1.71 cell line, downregulation of MUC1 in ESCC cell line TE-1 also remarkably reduced cells migration (data not shown). These results indicate that MUC1 promotes cell migration by upregulating MMP13.
Depletion of MUC1 reduces MMP13 expression and cell migration. (a) Cell lysates of human Ec1.71 cells stably expressing CTL-siRNA or MUC1-siRNA were collected and immunoblotted with antibody against MUC1-C and β-actin. (b) RT-PCR analysis on mRNA level of MMP13 and GAPDH control using total RNA from indicated cell lines. (c) The culture medium from indicated cells was collected. The activities of MMP13 were measured by gelatin-zymography (upper). Coomassie blue staining of SDS-PAGE without gelatin as loading control (lower). (d) Transwell assay were performed on the indicated cells (1 × 104; same as Figure 3c). (e) Transmigration cells were counted for each of the indicated cells. Columns, mean number of cells obtained in three independent experiments; bars, s.d.; ***P<0.001.
MUC1 Enhances Cell Metastatic Potential is Associated with Increased MMP13
To clarify the association of cell migration and MMP13, Ec1.71/MUC1-siRNA-B cells were transduced with pBabe-Flag-MMP13 or pBabe empty vector retroviruses, respectively. Immunoblotting of cell lysates from above cells showed that MMP13 expression was significantly increased in Ec1.71/MUC1-siRNA-B/pBabe-Flag-MMP13 cells but not in Ec1.71/MUC1-siRNA-B/pBabe vector cells (Figure 5a upper). Consistent with previous results, the migration rate of Ec1.71/MUC1-siRNA-B/pBabe-Flag-MMP13 cells was found to be significantly elevated when compared with that of Ec1.71/MUC1-siRNA-B/pBabe vector cells (P<0.005, Figure 5b and c). To investigate the effect of MUC1 on cell motile ability in more detail, and to address whether the effect correlates with MMP13, invasion, adhesion and aggregation assay were performed on Ec1.71/CTL-siRNA, Ec1.71/MUC1-siRNA, Ec1.71/MUC1-siRNA-B/pBabe vector and Ec1.71/MUC1-siRNA-B/pBabe-Flag-MMP13 cells. Invasion and adhesion were markedly reduced with MUC1 silence, but dramatically rescued when overexpression of MMP13 (Figure 5d and e). By contrast, the percentage of aggregated cells was about 33.3±3.8% in Ec1.71/CTL-siRNA cell (Figure 5f). Whereas it was increased to 45.8±3.8% with silencing of MUC1 in Ec1.71/MUC1-siRNA cells. No significant difference was found between Ec1.71/MUC1-siRNA and Ec1.71/MUC1-siRNA-B/pBabe vector cells. Interestingly, overexpression of MMP13 reduced the percentage of aggregated cells to 24.2±6.3%. These data revealed that the apparent function of MUC1 on cell metastatic potential is associated with MMP13.
MUC1-enhancing cell metastatic potential is associated with increased MMP13. (a) Ec1.71 MUC1-siRNA-B stable cell line was infected with pBabe-Flag-vector and pBabe-Flag-MMP13. The cell lysates were immunoblotted with MMP13, Flag and β-actin antibody. (b) Transwell assay were performed on the indicated cells (1 × 104; same as Figure 3c). Photomicrographs obtained at × 100 magnification. (c) Transmigration cells were counted for Ec1.71 MUC1-siRNA-B/Vector (open bar) and Ec1.71 MUC1-siRNA-B/MMP13 (solid bar). Columns, mean number of cells obtained in three independent experiments; bars, s.d. Invasion (d), adhesion (e) and aggregation assay (f) were performed on indicated cells. Columns, mean number of cells obtained in three independent experiments; bars, s.d.; *P<0.05, **P<0.01, ***P<0.001.
DISCUSSION
Overexpression of MUC1 and MMP13 is Correlated with Tumor Metastasis and Poor Outcome of Esophageal Cancer Patients
Esophageal carcinoma is one of the most aggressive tumors; its progression is relatively rapid and often manifested by lymph node metastasis. In our study, the patients with lymph node metastasis exhibited a significantly poorer prognosis than those without lymph node metastasis (P<0.01), consistent with the results from previous studies.14
MMP13 (collagenase-3) has a key role in extracellular matrix degradation and tumor metastasis and its overexpression was reported in ESCC.4 The expression of MUC1 in tumors may function as an anti-adhesion molecule that inhibits cell–cell aggregation, inducing dissemination of cells from tumor nests.15, 16 A high level of MUC1 expression was reported to be correlated with lymph node metastasis and associated with poor prognosis in ESCC patients, which implicate that MUC1 may serve as a novel biomarker for predicting lymph node metastasis and prognosis in ESCC.17 However, the mechanism in which manner MUC1 promotes ESCC metastasis remains unknown. In the present study, we demonstrate co-overexpression of MUC1 and MMP13 in ESCC patients, especially in patients with lymph node metastasis. Although we did not observe significant statistical correlation between overexpression of these two genes and poorer survival, which might be due to insufficient number of samples, the data demonstrated trends toward decreased survival rate in patients with overexpression of either one gene or both of them.
MUC1 Upregulates MMP13 Transcription in Cells
MUC1-C was explored to be able to interact with β-catenin and p53, thereby, co-activate transcription of the target genes.18, 19 The present investigation uncovers a strong correlation between the protein level of MUC1 and the mRNA level of MMP13 in tumor tissues. Further studies reveal that overexpression of MUC1-CD in 3Y1 cells is associated with an induction of MMP13 protein and mRNA. The promoter mapping experiments show that the highest activity locates within the P-402 promoter but not in the P-56 promoter, suggesting that there are stimulatory elements between –56 and –402 bp. Further study focused on the stimulatory elements of Runx-2 and Ets demonstrates that MUC1 promotes MMP13 transcription by Runx-2-binding site. Moreover, Ets regulates MMP13 by cooperating with Runx-2 when MUC1 is absent, however, Ets exerts dispensable on MMP13 expression in the presence of MUC1-CD.
Runx-2 is a transcription factor, which is required for IL-1 induction of MMP13. Increased expression of Runx-2 in the Brca1-deficiency mouse model induced esophagus tumorigenesis.20 How Runx-2 is involved in MMP13 regulation in human esophagus carcinoma is currently under investigation.
MUC1 Overexpression Promotes Cell Metastatic Potential Through Upregulating MMP13
Tumor metastasis consists of numerous consequent processes, which include migration, homotypic and heterotypic cell–cell adhesion, cell–matrix interaction, invasion into surrounding tissues, release from the primary tumor, intravasation, adhesion to vascular walls, extravasation and formation of new foci.21 Compiled studies have implicated MMPs in cell metastatic potential in physiological and pathological processes through the basis for the mechanism of ECM degradation. Tissue inhibitors of MMPs in general inhibit tumor cell invasion, angiogenesis and metastasis.22, 23, 24 Studies also shown that an elevated MMP13 expression resulted in an increased migration and survival of keratinocytes in vitro.25 The wound-healing assay demonstrated a significantly faster migration of 3Y1/MUC1-CD cells when compared with 3Y1/Vector cells, which is further corroborated by the transwell assay. Conversely, cell migration capacity significantly decreased when MUC1 expression was silenced by siRNA in Ec1.71 cells. Concomitantly to the loss of migration, MUC1 silencing in Ec1.71 cells induced markedly decreased invasion and adhesion. However, the cells favored more cell–cell aggregation with downregulation of MUC1. These results indicate that overexpression of MUC1 could facilitate the metastatic potential of tumor cells, which may contribute to tumor cell detachment from the primary tissue and promote the process of migration, invasion and metastasis. As expected, overexpression of MMP13 in MUC1 silenced Ec1.71 cells, completely rescued the cell motility and withdrew aggregation ability. Therefore, we conclude that MUC1 enhances cell metastastic capacity by increasing MMP13 gene expression.
In conclusion, our study finds that overexpression of MUC1 and MMP13 in ESCC patients is highly related to lymph node metastasis. We also elucidate the potential mechanism by which MUC1 stimulates MMP13 expression and thereby promotes cell metastasis. Our study suggests MUC1 as a novel diagnostic biomarker and therapeutic target for ESCC patients.
References
Enzinger PC, Mayer RJ . Esophageal cancer. N Engl J Med 2003;349:2241–2252.
Mori M, Barnard GF, Mimori K, et al. Overexpression of matrix metalloproteinase-7 mRNA in human colon carcinomas. Cancer 1995;75:1516–1519.
Deryugina EI, Quigley JP . Matrix metalloproteinases and tumor metastasis. Cancer Metastasis Rev 2006;25:9–34.
Etoh T, Inoue H, Yoshikawa Y, et al. Increased expression of collagenase-3 (MMP-13) and MT1-MMP in oesophageal cancer is related to cancer aggressiveness. Gut 2000;47:50–56.
Kufe D . Mucins in cancer: function, prognosis and therapy. Nat Rev Cancer 2009;9:874–885.
Sagara M, Yonezawa S, Nagata K, et al. Expression of mucin 1 (MUC1) in esophageal squamous cell carcinoma: its relationship with prognosis. Int J Cancer 1999;84:251–257.
Kufe D, Inghirami G, Abe M, et al. Differential reactivity of a novel monoclonal antibody (DF3) with human malignant versus benign breast tumors. Hybridoma 1984;3:223–232.
Huang L, Chen D, Liu D, et al. MUC1 oncoprotein blocks glycogen synthase kinase 3β–mediated phosphorylation and degradation of β-catenin. Cancer Res 2005;65:10413–10422.
Gao F, Yi J, Shi GY, et al. The sensitivity of digestive tract tumor cells to As2O3 is associated with the inherent cellular level of reactive oxygen species. World J Gastroenterol 2002;8:36–39.
Pendás AM, Balbín M, Llano E, et al. Structural analysis and promoter characterization of the human collagenase-3 gene (MMP13). Genomics 1997;40:222–233.
Chiu Y, Yang R, Hsieh K, et al. Stromal cell-derived factor-1 induces matrix metalloprotease-13 expression in human chondrocytes. Mol Pharmacol 2007;72:695–703.
Agata N, Ahmad R, Kawano T, et al. MUC1 oncoprotein blocks death receptor–mediated apoptosis by inhibiting recruitment of caspase-8. Cancer Res 2008;68:6136–6144.
Zhang J, Shen YL, Liu JW, et al. Antimetastatic effect of prodigiosin through inhibition of tumor invasion. Biochem Pharm 2005;69:407–414.
Shimada Y, Imamura M, Watanabe G, et al. Prognostic factors of oesophageal squamous cell carcinoma from the perspective of molecular biology. Br J Cancer 1999;80:1281–1288.
Ligtenberg MJ, Buijs F, Vos HL, et al. Suppression of cellular aggregation by high levels of episialin. Cancer Res 1992;52:2318–2324.
Makiguchi Y, Hinoda Y, Imai K . Effect of MUC1 mucin, an anti-adhesion molecule, on tumor cell growth. Jpn J Cancer Res 1996;87:505–511.
Song ZB, Gao SS, Yi XN, et al. Expression of MUC1 in esophageal squamous-cell carcinoma and its relationship with prognosis of patients from Linzhou city, a high incidence area of northern China. World J Gastroenterol 2003;9:404–407.
Huang L, Ren J, Chen D, et al. MUC1 cytoplasmic domain coactivates Wnt target gene transcription and confers transformation. Cancer Biol Ther 2003;2:702–706.
Wei X, Xu H, Kufe D . Human MUC1 oncoprotein regulates p53-responsive gene transcription in the genotoxic stress response. Cancer Cell 2005;7:167–178.
Cao L, Xu X, Cao LL, et al. Absence of full-length Brca1 sensitizes mice to oxidative stress and carcinogen-induced tumorigenesis in the esophagus and forestomach. Carcinogenesis 2007;28:1401–1407.
Chen NH, Liu JW, Zhong JJ . Ganoderic acid Me inhibits tumor invasion through down-regulating matrix metalloproteinases 2/9 gene expression. J Pharmacol Sci 2008;108:212–216.
Chen NH, Liu JW, Zhong JJ . Ganoderic acid T inhibits tumor invasion in vitro and in vivo through inhibition of MMP expression. Pharmacol Rep 2010;62:150–163.
Vanhoecke BW, Bracke ME, Kloosterboer HJ, et al. Tibolone and its metabolites inhibit invasion of human mammary carcinoma cells in vitro. Maturitas 2006;54:229–237.
Sancéau J, Truchet S, Bauvois B . Matrix metalloproteinase-9 silencing by RNA interference triggers the migratory-adhesive switch in Ewing's sarcoma cells. J Biol Chem 2003;278:36537–36546.
Uitto VJ, Baillie D, Wu Q, et al. Fusobacterium nucleatum increases collagenase 3 production and migration of epithelial cells. Infect Immun 2005;73:1171–1179.
Acknowledgements
This work was supported by program 30871363, 81071666 by NNSF of China, program NCET-08-0349 and the SRF for ROCS by State Education Ministry, the program 07pj14065 and 06ZR14067 by Science and Technology Commission of Shanghai Municipality and program E03003 and J50201 by the Shanghai Municipal Education Commission. The authors are very grateful to Dr Donald Kufe (Dana-Farber Cancer Institute, Harvard Medical School) for expert advice and to Dr Zhimin Yuan (UT Health Science Center) for critical comments on the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Laboratory Investigation website
Supplementary information
Rights and permissions
About this article
Cite this article
Ye, Q., Yan, Z., Liao, X. et al. MUC1 induces metastasis in esophageal squamous cell carcinoma by upregulating matrix metalloproteinase 13. Lab Invest 91, 778–787 (2011). https://doi.org/10.1038/labinvest.2011.12
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/labinvest.2011.12
Keywords
This article is cited by
-
Analysis of Molecular Genetic Variants of Lgals4 in Esophageal Cancer: A Preliminary Report
Biochemical Genetics (2024)
-
Crosstalk between MMP-13, CD44, and TWIST1 and its role in regulation of EMT in patients with esophageal squamous cell carcinoma
Molecular and Cellular Biochemistry (2021)
-
CircWHSC1 promotes ovarian cancer progression by regulating MUC1 and hTERT through sponging miR-145 and miR-1182
Journal of Experimental & Clinical Cancer Research (2019)
-
Clinic implication of MUC1 O-glycosylation and C1GALT1 in esophagus squamous cell carcinoma
Science China Life Sciences (2018)
-
MUC1 induces acquired chemoresistance by upregulating ABCB1 in EGFR-dependent manner
Cell Death & Disease (2017)