Abstract
MicroRNAs are a class of recently discovered small RNA molecules that regulate other genes in the human genome. Studies in human cells and model organisms have begun to reveal the mechanisms of microRNA activity, and the wide range of normal physiological functions they influence. Their alteration in pathologic states from cancer to cardiovascular disease is also increasingly clear. A review of current evidence for the role of these molecules in human health and disease will be helpful to pathologists and medical researchers as the fascinating story of these small regulators continues to unfold.
Similar content being viewed by others
Main
Modern medicine has begun to gather an understanding of the molecular mechanisms that give rise to the cellular patterns long seen through the pathologist's microscope. The ‘central dogma’ of DNA genes copied to messenger RNA transcripts that guide the synthesis of proteins still provides the basic framework connecting genetics to medical phenomena. Yet, as is often the case in biology and medicine, the details and exceptions can be of particular interest.
The recently discovered tiny RNA molecules known as microRNAs are important regulators of the other genes in the human genome. Our understanding of these small RNA molecules is still in its infancy, but examples from human cells as well as mice and other model organisms indicate that microRNA-mediated gene regulation is likely to play key roles in human development, cellular differentiation, adaptation to the environment, oncogenesis, and host cell interactions with pathogens. For pathologists, methods for measuring microRNA levels offer a new set of tools that may be of use in improving our understanding and classification of many diseases.
DISCOVERY OF SMALL RNA
Previous decades unveiled a number of important cellular roles for RNA distinct from the information-encoding transcript of messenger RNA. Ribosomal RNA is both a structural and catalytic part of the ribosome;1 tRNAs play an informational role in decoding the codons of the messenger RNA while carrying amino acid residues for protein synthesis;2 the small nuclear RNAs of the spliceosome recognize splice sites and catalyze alternate splicing of pre-mRNA transcripts;3 and several small nucleolar RNAs are involved in covalent modification of other RNA bases.4 To continue the list, the 7SL RNA is a part of the signal recognition particle that channels proteins into the endoplasmic reticulum during synthesis;5 the telomerase complex carries its own RNA as a template for extending the ends of chromosomes;6 and the very long noncoding RNA XIST is required for X-chromosome silencing and dosage compensation in human females.7
The discovery of very short noncoding RNAs as regulators of gene activity began almost 15 years ago when researchers studying larval development in the nematode worm C. elegans discovered a gene, lin-4, that encoded short RNA transcripts able to inhibit translation of the messenger RNA of a different gene, lin-14, by binding to its 3′ untranslated region.8 In hindsight, lin-4 was the first of the microRNA class of genes to be characterized. We now realize that there are hundreds of similar microRNA genes in the human genome. However, before the widespread importance of microRNAs came to be appreciated, another kind of small RNA molecule took center stage.
Geneticists studying pigment production in petunias in the late 1980s made the paradoxical observation that plants carrying extra copies of a pigment production transgene sometimes produced flowers with patchy pigment, or little pigment at all.9, 10 The mechanism for this trans-acting ‘cosuppression’ silencing of the endogenous pigment production gene was not fully known, but RNA produced by the transgene array was a prime suspect as a mediator of silencing.
Shortly thereafter, researchers studying the inconsistent ability of injected RNA to silence genes in C. elegans tested whether sense-, anti-sense, or double-stranded RNA homologous to a gene was the most active in suppressing endogenous gene activity. Surprisingly, double-stranded RNA had the greatest silencing effect.11 Further investigations by several labs revealed that short (21–22 nucleotide) double-stranded RNA cassettes (short interfering RNA, or siRNA) were generated from longer double-stranded RNA by an enzymatic system within the cell, and functioned by causing cleavage of the endogenous gene mRNA to which they were homologous.12, 13, 14, 15 Although human cells have not yet been shown to generate endogenous siRNAs, the evolutionarily conserved enzymatic machinery used with microRNAs can recognize siRNAs. As a result, exogenous siRNA reagents can be used to ‘knock down’ human gene function for research, and potentially for therapeutic purposes.
Spurred on by these findings, a large number of other researchers have begun to flesh out the picture of small RNA biology in human cells. Identification of short RNA molecules from a variety of healthy and diseased human tissues indicates that there may be well over 500 distinct microRNAs in the human genome.16, 17 Some of the roles of these microRNAs in normal human biology and in disease are beginning to come into focus.
Notably, entirely new classes of small RNA molecules such as the PIWI-associated RNAs (piRNAs) in the male germline indicate that even the broad outlines of this subject have not yet been fully delineated.18 Results from the latest generations of genome-tiling microarrays and high-throughput sequencing indicate that there may be a considerably richer universe of non-coding RNAs of various sizes, produced from many different regions of the genome, than had been previously supposed.19, 20, 21 In addition, comparisons between animals with increasingly complicated bodies have been used to suggest that the proportion of non-coding RNA to protein-coding RNA is correlated with developmental complexity.22, 23, 24
MicroRNAs
Mature microRNAs are single-stranded RNA molecules that are approximately 21 or 22 nucleotides long. Most microRNAs are generated from primary transcripts (pri-miRNAs) produced by RNA polymerase II, the same RNA polymerase that transcribes protein-coding genes, but some microRNAs in repetitive regions of the genome are transcribed by RNA polymerase III.25, 26, 27 Some microRNA primary transcripts encode only a single mature microRNA, while other loci contain clusters of microRNAs that appear to be produced from a single primary transcript. Approximately half of all human microRNA genes are contained within the introns of protein-coding genes, while others reside apart from known genes, or in the exons of untranslated genes.28 Control of pri-miRNA synthesis is incompletely understood, but some of the same transcription factors that regulate protein-coding genes can apparently also regulate pri-miRNAs.29
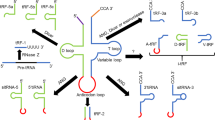
Within the primary transcript, the microRNA-encoding portions form imperfect stem-loop hairpin structures. A nuclear protein complex called the ‘Microprocessor,’ consisting of the Drosha type III RNase, the double-stranded RNA-binding protein DGCR8, and a number of other proteins, cleaves the hairpin-loop pre-miRNAs away from the rest of the primary transcript, permitting export of pre-miRNAs through the nuclear pore to the cytoplasm.30 There, the pre-miRNAs are further processed by a protein complex containing the Dicer type III RNase that cleaves the microRNA to its mature size.31, 32 In general, only one strand of the original precursor is kept as a mature microRNA. The mature microRNA becomes associated with a final protein complex called the miRISC (microRNA-induced silencing complex), which typically contains an Argonaute family protein, and which carries out the silencing of the genes targeted by the microRNA (Figure 1). Within many miRNA sequences, the so-called 5′ ‘seed’ region (from base 2 to base 8 or 9) is particularly important for target site recognition, but sequence context and base pairing between regions in the 3′ half of the microRNA and the target site can also contribute to the function of the microRNA.33, 34, 35
It appears that at least in some cells, the processing steps required to generate mature microRNAs from pri-miRNA transcripts are actively regulated, which may give extra flexibility for controlling the amount of active microRNA in the cell.36
Mechanisms of MicroRNA Action
Unlike siRNAs, which require near-perfect complementarity to the messenger RNA they target for cleavage, most microRNAs form imperfectly complementary stem-loop structures, pair imperfectly with sites in the 3′ untranslated region (UTR) of their target mRNAs, and can apparently act by decreasing target messenger RNA levels or by directly inhibiting translation.37, 38, 39, 40 In some cases microRNAs have been proposed to act relatively promiscuously and to inhibit multiple targets.38, 41 Many genes have predicted target sites for several different microRNAs in their 3′ untranslated regions, indicating the possibility for combinatorial action of multiple microRNAs to maximize inhibition of gene activity, although for most genes these predictions have not yet been tested. Preliminary in silico estimates are that about one-third of all protein-coding genes in the human genome may be regulated in part by microRNAs.35, 38 The prediction of target genes for particular miRNAs is a difficult computational challenge, and is impaired by the lack of data sets describing alterations in individual protein levels in the proteome in response to alterations in miRNA levels. Target predictions based on mRNA downregulation data are more advanced, due to the greater tractability of mRNA expression profiling. When more protein data are available, it will be interesting to compare the microRNA-target interactions that correlate with effects on target mRNA levels and protein levels.
Biochemical experiments have begun to examine microRNA interactions with their target genes. Cellular subfractionation studies indicate that many miRNAs are found in the polyribosome fraction, which would be most consistent with a role in decreasing productive extension of the polypeptide chain after initiation of translation.42, 43 Other proposed mechanisms of miRNA action include interference with recognition of the 5′ cap of the mRNA to prevent translation initiation, destabilization of mRNA via deadenylation of the poly-A tail, and recruitment of the ribosome-inhibitory protein eIF6.44, 45, 46, 47 The detailed mechanisms of miRNA activity, including a full list of the protein components of the active miRISC complex for various miRNAs, await further elucidation. A recent paper underscores this point with the surprising observation that under some conditions, such as cell cycle arrest, microRNAs can upregulate translation of a target protein.48
Evolutionary Considerations
Comparisons between humans and other animals show that many microRNAs are well conserved in evolution, and are likely part of important ancient pathways of gene regulation, while other microRNAs are more restricted in their phylogenetic distribution.23, 24, 49 A comparison of brain-derived microRNAs in humans and chimpanzees uncovered a large number of novel primate- and human-specific microRNAs that have been proposed to relate to the increased complexity and specialization of brain development that occurred in human evolution.17
Many microRNAs in humans belong to families whose members reside at different chromosomal loci, but are very similar in sequence. Similar to protein-coding gene families with many closely related members, there may be functional redundancy or partial redundancy of these microRNAs. This redundancy will increase the challenge of discovering the microRNA-target relationships and their functional consequences. For example, there are 11 distinct members of the let-7 microRNA family in humans, differing from each other by a few bases at most.50
The short length of microRNAs and their binding sites within target genes should give them the opportunity to mutate and change specificity rapidly in evolution. A striking example of the potent effect of such mutation comes from Texel sheep, an exceptionally muscular breed, where a genetic quantitative trait locus has been mapped to a mutated site in the myostatin gene 3′UTR. The mutation creates a new target site for the miR-1 and miR-206 microRNAs which are abundantly expressed in skeletal muscle, causing downregulation of the skeletal muscle-inhibitory myostatin protein and giving rise to the muscle hypertrophy phenotype.51 The resultant phenotype is similar to the hyper-muscularity resulting from coding region or splice site mutations in the myostatin gene reported in cattle, mice, dogs, and humans.52, 53 In humans, some evidence for a similar story has emerged for a hypertension-associated allele of the angiotensin receptor AGTR1 gene: the 1166C allele disrupts a miR-155 target site in the 3′UTR of the gene, potentially preventing downregulation of the AGTR1 protein.54
Overall, it appears that there is a relative depletion of predicted target sites for microRNAs in the 3′ UTRs of messenger RNAs that are coexpressed in the same cells or tissues as the microRNAs, indicating that microRNA-based selective pressures may have a substantial effect on the evolution of potential target genes.55 In a different arena of selective pressure, the capacity for rapid evolutionary change in microRNA genes and their target sites has been hypothesized as providing an advantage to animals in responding to rapidly evolving parasites and pathogens.56
MicroRNAs IN DEVELOPMENT AND PHYSIOLOGY
Profiles of miRNAs in Normal Human Tissues and Cells
Cloning and measurement of microRNAs from various human tissues has proceeded quickly. We now have an overview of the sites of expression of many microRNAs.57 About one-third of microRNAs show substantial tissue specificity, while the others may vary in expression level but are not particularly tissue or cell type-specific, at least at the resolution of currently available data. Among the most specific miRNA expression patterns are that of miR-122 in the liver, miR-375 in pancreatic islet tissue, miR-142 and miR-223 in the hematopoietic system, and miR-1 and miR-133 in muscle.57, 58, 59 The utility of some tissue-specific microRNAs for identifying the tissue of origin of poorly differentiated metastases of unknown primary tumors in human patients has been shown in a proof-of-concept analysis.60 Reports of the phenotypes of knockout mice for different microRNA genes and tissue-specific Dicer mutants have begun to appear in the literature, giving valuable insights into the cellular processes that become derailed when particular microRNAs are absent. A cautionary note concerning the overall importance for microRNAs in animal development may be sounded by the results of Dicer mutation in zebrafish, where many cell lineages and embryologic structures developed adequately in the absence of known microRNA pathways.61 It may be that some animal tissues and cell lineages will be more dependent on microRNA regulation than others.
Stem Cell Biology
A small number of microRNAs are expressed mainly in embryonic stem (ES) cells, and attention has naturally focused on their roles in maintaining the pleuripotent undifferentiated state, self-renewal, and other characteristics of ES cells.62, 63 Some evidence for such roles comes from the poor proliferation and differentiation potential of Dicer-null murine ES cells, and from the small number of germline stem cells produced in Drosophila mutants of Dicer1 or its accompanying RNA-binding protein, Loquacious.64, 65, 66, 67 In contrast, mouse ES cells null for the microprocessor complex member DGCR8, which is also required for microRNA biogenesis, showed a less severe impairment of division in culture while still being unable to differentiate normally into other cell types when appropriately stimulated.68 Together, the mammalian ES cell studies of Dicer and DGCR8 mutants show these proteins having overlapping but not identical functions, suggesting that one or both may be involved in other pathways besides the microRNA pathway as it is currently understood. Targeted disruption of ES cell-specific microRNAs can be expected to shed further light on exactly which miRNA effectors may contribute to the observed phenotypes.
Germline
Tissue-specific ablation of the Dicer gene in mouse oocytes results in abnormalities culminating in meiosis I arrest with severe defects in meiotic spindle formation and chromosome organization. In addition, mRNAs that are normally degraded in meiotic maturation in oocytes, and that have predicted target sites for oocyte-expressed microRNAs, were present at elevated levels in the Dicer-null oocytes, suggesting that the microRNAs may participate in specifying their destruction.69 This is reminiscent of the situation shortly after fertilization in zebrafish zygotes, where a single microRNA, miR-430 is postulated to destabilize hundreds of maternally-expressed mRNAs that would otherwise linger in the zygote. In support of this, zebrafish Dicer mutants have increased zygotic levels of mRNAs with predicted miR-430 target sites.70
Skeletal and Cardiac Muscle
The human and mouse genomes each contain two bicistronic loci, each of which expresses miR-1 and miR-133 microRNAs in skeletal and cardiac muscle.71, 72 Selective disruption of the miR-1 gene (miR-1-2) from one of the two loci in mice revealed that the absence of this microRNA causes 50% embryonic lethality with frequent ventricular septal defects. Among miR-1-2 knockout mice who survived to adulthood, abnormal cardiac electrical conduction and hyperplasia of cardiac myocytes was observed, despite grossly normal cardiac anatomy and function.73 The results from this mouse strain, which, it should be emphasized, only inactivates one of the two miR-1 microRNAs known to be expressed in cardiac muscle, may indicate an important role for mir-1-2 in the embryological development of the heart, and a lesser role in normal adult cardiac tissue. Of course, the degree of compensation provided by the other miR-1 locus will need to be examined to get a deeper understanding of the role of this microRNA family in cardiac development and maintenance. In another recent study, interference with the muscle-specific miR-133 family members using antagomir (essentially, antisense oligonucleotide) techniques caused the development of cardiac hypertrophy in mice, indicating that the normal role of this cardiac microRNA may be to prevent undesirable myocyte hypertrophy.74
Hematopoiesis and Immunity
Normal hematopoiesis and immunity are guided by microRNA-mediated gene regulation. miR-181 is expressed in hematopoietic tissues, and at lower levels in several other tissues such as muscle.75 Overexpression of miR-181 in murine hematopoietic progenitor cells increases the relative proportion of B cells to T cells in the peripheral blood; interpretation of the mechanism of this effect is complicated by the role played by this microRNA in modulating thymocyte T cell receptor signalling and clonal selection, but it clearly influences the final balance of the two major lymphoid lineages.76, 77, 78
Immunological studies in mice have shown that miR-155 is expressed in germinal center B cells, and is required for normal germinal center formation, antibody titers in response to antigen, and plasma cell differentiation or persistence.79, 80, 81 Normal functioning of T lymphocytes and dendritic cells also appears to depend on this microRNA.79, 80 In the absence of miR-155, helper T cells deviate toward the TH2 phenotype, producing increased levels of IL-4 and less interferon gamma than controls.79, 80
In both mouse and human cells, the earliest stages of erythroid cell differentiation are accompanied by dramatic upregulation of miR-451.82, 83 Indeed, even peripheral red blood cell preparations show significant amounts of miR-451, perhaps indicating a role for this microRNA in the final regulation of mRNA in reticulocytes.84
Nervous System
miR-9, miR-124, and miR-128 are among the microRNAs most highly and specifically expressed in the mammalian brain.85, 86 Deletion of the well-conserved miR-9 in fruit flies causes duplication of sensory neurons and sensory organs in development.87 The effects of overexpression or temporarily knocking down miR-124 in chick neural tubes, in contrast, are somewhat controversial, and show, at most, a modest impact of this microRNA on neuronal differentiation.88, 89 Neuronal differentiation in neuroblastoma and embryonal carcinoma cell lines in vitro is enhanced by miR-124, at least in part due to miR-124 inhibiting expression of a regulator of alternate mRNA splicing, PTBP1.90 Given the apparent rich diversity of microRNAs expressed at low levels in the mammalian CNS, subtle phenotypes or highly cell type-specific microRNA roles may be the rule in this organ.17 A hint of these possibilities comes from an example in the nematode worm C. elegans, where left–right asymmetry of chemosensory receptor expression in some neurons is controlled by a microRNA named lsy-6.91
MICRORNA IN DISEASE STATES
Given their important roles in normal physiology, it is no surprise that microRNAs have come under close scrutiny in a variety of diseases. MicroRNAs have been shown to play contributory roles in cancer, heart disease, infectious disease, and other medical conditions.
Cancer
Some miRNAs appear to act as oncogenes, by contributing to the transformed phenotype when expressed at elevated levels in cancers. miR-21 is relatively overexpressed in glioblastoma multiforme, cervical cancer, breast cancer, and many other solid tumor types.92, 93, 94 Increased expression of miR-21 appears to contribute to decreased apoptosis in the malignant cells.93 Similarly, the locus containing seven microRNAs of the miR-17-92 polycistron cluster at human chromosome 13q31 is amplified in a variety of B cell lymphomas, nasal-type NK/T cell lymphoma, and solid tumors.95 Overexpression of a portion of this polycistron, miR-17-19b, in a mouse lymphoma model where the c-myc gene is regulated by the immunoglobulin heavy chain enhancer, showed cooperation between miR-17-19b and c-myc in oncogenesis, causing more rapid lymphoma growth and shorter animal survival.96 The targets and mechanisms of miR-17-92 action have yet to be elucidated, but Myc-regulated expression of this polycistron also appears to determine the angiogenic potential of colon adenocarcinomas in a different mouse model system.97
Conversely, some microRNAs can act as tumor suppressors, genes whose deletion or mutation helps cells along the multi-step process of tumorigenesis. Circumstantial evidence for this possibility first emerged in early microRNA profiling experiments where tumor samples appeared to have lower overall levels of microRNA expression than normal tissue samples, both in human clinical material and tissues from murine cancer models.60 There is some debate at present concerning the extent of global miRNA suppression in human cancers; some of the differences in reported findings may relate to choices of normal samples for comparison, different analytic platforms, a lack of internal standards for each analyte measured, or the use of normalization procedures that could minimize actual global differences between tumor and normal samples.60, 92
Some evidence supporting a role for global miRNA suppression in cancer cells came from a relatively small series of non-small cell lung cancer samples that showed decreased survival of patients whose tumors had lower levels of Dicer mRNA expression.98 A helpful recent study used RNA interference methods to globally ‘knock down’ mature microRNA levels by inhibiting expression of Dicer1, Drosha, or DGCR8.99 Impaired miRNA pathways led to increased growth of mouse and human cancer cell lines in soft agar cultures, and more malignant behavior when cells were injected into nude mice. Deletion of one or both copies of the Dicer1 gene was also shown to lead to greater tumor burden in a Kras point-mutant mouse model of lung cancer, further supporting the idea that small RNA pathways dependent on Dicer tend to act as an overall brake on cancer growth.99 Overexpression of let-7 g reversed the in vitro effects of Dicer1 knockdown, indicating that let-7 family members may be key tumor suppressors among microRNAs. These findings fit well with earlier mechanistic studies of the let-7 miRNA family that identified Ras genes as targets of inhibition by let-7 miRNAs in both nematodes and human cancer cells.100
Other examples of miRNA involvement in cancer are suggestive but less easily classified. An initial report described overexpression of miR-155 as a hallmark of pediatric Burkitt's lymphoma, but a subsequent study found that miR-155 was only overexpressed in a minor subset of Burkitt's lymphoma cells, those exhibiting Epstein–Barr virus latency pattern III.101, 102 Studies of the 13q14 locus commonly deleted in chronic lymphocytic leukemia (CLL) indicated that miR-15a and miR-16-1, the two microRNAs at the locus, could act as tumor suppressors, perhaps by downregulating the antiapoptotic gene BCL2.103, 104, 105 The New Zealand Black mouse model for CLL has also been shown to have a sequence variant in the miR-16 gene Mirn16-1 that causes a modest decrease in levels of miR-16. Attempts to use the overall profile of microRNA expression to predict patient outcome in CLL have identified correlations, but these did not provide extra information beyond conventional prognostic markers such as IgVH gene hypermutation.104 More recent work measuring miRNA expression in CLL using direct miRNA cloning and quantitative RT-PCR methods instead of microarray hybridization found no correlation between miR-15a or miR-16-1 expression and Bcl2 levels.106 Hopefully, further study can clarify the reasons for these different results, and perhaps give further insight into the reasons why the 13q14 deletion is, in fact, a marker for good prognostic in CLL.
MicroRNAs now seem likely to be important downstream target genes of previously identified transcription factors, as well. Four recent studies identified miR-34a as a gene regulated by the tumor suppressor p53 transcription factor. These reports provide evidence that miR-34a is important for p53-mediated apoptosis, and possibly for other p53-mediated responses to cellular stress.107, 108, 109, 110 The mechanism of these effects has yet to be established.
Finally, tumor phenotypes associated with extensive disease, such as invasion of surrounding tissues and metastasis, are also under the influence of microRNA controls. A new cellular pathway governing tumor invasiveness and metastasis has been mapped out in a breast cancer model system in which human breast cancer cells are implanted into the mammary fat pads of mice. The transcription factor Twist regulates expression of microRNA miR-10b, which inhibits translation of the homeobox gene HOXD10, preventing it from repressing the gene for the GTPase RHOC, which has been shown to be a metastasis-promoting protein in several studies.111, 112 The net effect of increased expression of miR-10b is increased invasiveness and metastasis of the tumor cells, a finding consistent with increased expression of this microRNA in primary tumors from patients who had metastatic disease.111
On the more clinical end of the research spectrum, profiling of microRNAs in many other kinds of cancers is proceeding rapidly, with recent examples from cervical, prostate, pancreatic, and germ cell tumors, among many others.92, 94, 113, 114, 115, 116, 117, 118 Characteristic microRNA patterns offer new information that could yield advances in tumor classification and prognosis determination, although the clinical validation of such efforts will require some time.
Heart Disease
Several recent papers have described the contribution of microRNAs to the physiology of cardiac hypertrophy under conditions of stress and heart failure. Exogenous overexpression of some miRNAs that are upregulated in failing hearts is sufficient to cause cardiac myocyte hypertrophy in culture, and transgenic overexpression of miR-195 in a mouse's heart causes frank cardiac hypertrophy.119, 120 These experiments demonstrate the potent effect of microRNAs in modulating the physiology of complex organs in the whole animal. In contrast, knocking out another miRNA with a cardiac expression pattern, miR-208, showed that it is necessary for the cardiac hypertrophy and upregulation of beta-myosin heavy chain that occurs in response to increased cardiac afterload.119 The expression of miR-208 was also required for the cardiac upregulation of beta-myosin heavy chain that occurs in the absence of normal thyroid hormone signaling, showing that microRNAs can play roles in the endocrine coordination of cardiac responses.119
Infectious Disease
A number of viruses have long been known to express short non-coding transcripts from their genomes, and it now seems that some short non-coding viral RNAs may make use of, or subvert, the machinery of the microRNA pathway in the host cell. The large γ herpesvirus Epstein–Barr virus appears to encode two clusters of microRNAs, one cluster in the introns of the BART gene, another in the 3′ and 5′ untranslated regions of the BHRF1 gene.121, 122, 123 The numerous predicted targets for these viral miRNAs include genes involved in cell proliferation, apoptosis, and immune signaling.123 Similarly, other herpesviruses including Kaposi's sarcoma virus, cytomegalovirus, as well as the SV40 DNA tumor virus, have all been reported to express microRNAs from their genomes.122, 124, 125, 126, 127, 128 The single SV40 miRNA is necessary for regulating the levels of early genes in the viral life-cycle, and contributes to virally-infected cell resistance to cytoxic T cell lysis.126 In contrast, initial analyses of some viruses with RNA genomes, such as yellow fever virus, hepatitis C virus (HCV), and human immunodeficiency virus 1 (HIV) have so far failed to detect virus-encoded microRNAs.122 Interestingly, HCV makes use of the host hepatocyte-expressed miR-122 to increase the replication of its own RNA genome.129
Some viruses seem to block activity of miRNA pathways altogether. Replication of the primate foamy virus is inhibited by the host cell miR-32, but the virus expresses a transcriptional activator protein, Tas, that can interfere with miRNA activity.130 Other inhibitors of RNA-mediated gene silencing pathways have been reported in influenza A, vaccinia, HCV, HIV-1, and Ebola viruses. The specificity of these effects and their role in productive viral infection certainly warrant further investigation.131, 132, 133, 134
Efforts are also underway to determine which host cell miRNAs are upregulated in response to individual pathogens. For the moment, miR-155 appears to be a major part of the macrophage inflammatory response, as it is upregulated in murine bone marrow macrophages by Toll-like receptor ligands such as polyriboinosinic: polyribocytosinic acid (poly-I:C), lipopolysaccharide, hypomethylated DNA, and the synthetic lipoprotein Pam3CSK4, as well as interferon beta.135 Mice homozygous for an unusual hypomorph allele of Dicer1 show increased susceptibility to infection by vesicular stomatitis virus (VSV).136 VSV mutated to ablate putative binding sites for host miR-24 and miR-93 has increased virulence, suggesting that these host microRNAs could work in a Dicer1-dependent manner to limit the course of the viral infection.136 In another twist, several cellular microRNAs have been shown to bind HIV-1 encoded transcripts, limiting production of the viral proteins, but ironically contributing to the ability of the retrovirus to remain latent and undetected by the immune system in quiescent CD4-positive T cells.137
PRACTICAL ASPECTS
A number of recent methodological papers have provided detailed protocols for measuring microRNA levels in different kinds of specimens. Real time PCR, microarrays, Luminex bead arrays, Northern blotting, and direct cloning and sequencing approaches have all been applied with success, although there may be some systematic differences between some methods.60, 138, 139, 140, 141, 142, 143, 144 In situ hybridization detection of microRNAs has also been reported by several groups.145, 146, 147 It is noteworthy that standard formalin-fixation and paraffin-embedding of tissues seems to preserve microRNAs in a reasonably intact and extractable state.146, 148, 149 Once these tools are clinically validated more extensively, they should enable microRNA measurements to become a part of the pathologist's diagnostic armamentarium.
CONCLUSIONS
MicroRNAs are important functional threads in the cellular tapestry seen through the pathologist's microscope. Their normal activities help to coordinate multicellular life, and their dysregulation can contribute to a variety of diseases. The functional studies described here are only the first forays into better understanding of microRNA roles. Further studies correlating microRNA activity with mRNA and protein levels in clinically relevant specimens, and more detailed mechanistic studies using cell- and tissue-specific manipulation of microRNA levels in animal models will likely reveal new surprises. Although there is no reason to believe that these small RNA molecules are more important than transcription factors and other protein-encoded gene products in the overall life of the cell, they represent a significant component of the apparatus that regulates gene activity. We can speculate that these small strands will help us to make many meaningful new biological and medical connections in coming years.
NOTE
Space restrictions prevented the discussion or citation of the many other contributions made by numerous labs in this rapidly progressing field. I thank Andrew Fire, the members of the Fire lab at Stanford, and my pathology department clinical colleagues for helpful comments and critical review of the article.
References
Beringer M, Rodnina MV . The ribosomal peptidyl transferase. Mol Cell 2007;26:311–321.
Soll D, RajBhandary UL . The genetic code—thawing the ‘frozen accident’. J Biosci 2006;31:459–463.
Padgett RA, Grabowski PJ, Konarska MM, et al. Splicing of messenger RNA precursors. Annu Rev Biochem 1986;55:1119–1150.
Smith CM, Steitz JA . Sno storm in the nucleolus: new roles for myriad small RNPs. Cell 1997;89:669–672.
Egea PF, Stroud RM, Walter P . Targeting proteins to membranes: structure of the signal recognition particle. Curr Opin Struct Biol 2005;15:213–220.
Theimer CA, Feigon J . Structure and function of telomerase RNA. Curr Opin Struct Biol 2006;16:307–318.
Masui O, Heard E . RNA and protein actors in X-chromosome inactivation. Cold Spring Harb Symp Quant Biol 2006;71:419–428.
Lee RC, Feinbaum RL, Ambros V . The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993;75:843–854.
van der Krol AR, Mur LA, Beld M, et al. Flavonoid genes in petunia: addition of a limited number of gene copies may lead to a suppression of gene expression. Plant Cell 1990;2:291–299.
Napoli C, Lemieux C, Jorgensen R . Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell 1990;2:279–289.
Fire A, Xu S, Montgomery MK, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998;391:806–811.
Hammond SM, Bernstein E, Beach D, et al. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 2000;404:293–296.
Zamore PD, Tuschl T, Sharp PA, et al. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21–23 nucleotide intervals. Cell 2000;101:25–33.
Tuschl T, Zamore PD, Lehmann R, et al. Targeted mRNA degradation by double-stranded RNA in vitro. Genes Dev 1999;13:3191–3197.
Hamilton AJ, Baulcombe DC . A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 1999;286:950–952.
Bentwich I, Avniel A, Karov Y, et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet 2005;37:766–770.
Berezikov E, Thuemmler F, van Laake LW, et al. Diversity of microRNAs in human and chimpanzee brain. Nat Genet 2006;38:1375–1377.
Girard A, Sachidanandam R, Hannon GJ, et al. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 2006;442:199–202.
Bertone P, Stolc V, Royce TE, et al. Global identification of human transcribed sequences with genome tiling arrays. Science 2004;306:2242–2246.
Jongeneel CV, Delorenzi M, Iseli C, et al. An atlas of human gene expression from massively parallel signature sequencing (MPSS). Genome Res 2005;15:1007–1014.
Cheng J, Kapranov P, Drenkow J, et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science 2005;308:1149–1154.
Mattick JS, Makunin IV . Non-coding RNA. Hum Mol Genet 2006;15 (Spec No 1):R17–R29.
Sempere LF, Cole CN, McPeek MA, et al. The phylogenetic distribution of metazoan microRNAs: insights into evolutionary complexity and constraint. J Exp Zoolog B Mol Dev Evol 2006;306:575–588.
Hertel J, Lindemeyer M, Missal K, et al. The expansion of the metazoan microRNA repertoire. BMC Genomics 2006;7:25.
Bracht J, Hunter S, Eachus R, et al. Trans-splicing and polyadenylation of let-7 microRNA primary transcripts. RNA 2004;10:1586–1594.
Lee Y, Kim M, Han J, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J 2004;23:4051–4060.
Borchert GM, Lanier W, Davidson BL . RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol 2006;13:1097–1101.
Rodriguez A, Griffiths-Jones S, Ashurst JL, et al. Identification of mammalian microRNA host genes and transcription units. Genome Res 2004;14:1902–1910.
Woods K, Thomson JM, Hammond SM . Direct regulation of an oncogenic micro-RNA cluster by E2F transcription factors. J Biol Chem 2007;282:2130–2134.
Yi R, Qin Y, Macara IG, et al. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev 2003;17:3011–3016.
Ketting RF, Fischer SE, Bernstein E, et al. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev 2001;15:2654–2659.
Hutvagner G, McLachlan J, Pasquinelli AE, et al. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 2001;293:834–838.
Vella MC, Choi EY, Lin SY, et al. The C. elegans microRNA let-7 binds to imperfect let-7 complementary sites from the lin-41 3′UTR. Genes Dev 2004;18:132–137.
Grimson A, Farh KK, Johnston WK, et al. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell 2007;27:91–105.
Lewis BP, Burge CB, Bartel DP . Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005;120:15–20.
Thomson JM, Newman M, Parker JS, et al. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev 2006;20:2202–2207.
Doench JG, Sharp PA . Specificity of microRNA target selection in translational repression. Genes Dev 2004;18:504–511.
Lim LP, Lau NC, Garrett-Engele P, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 2005;433:769–773.
Wightman B, Ha I, Ruvkun G . Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 1993;75:855–862.
Brennecke J, Hipfner DR, Stark A, et al. Bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 2003;113:25–36.
Krutzfeldt J, Rajewsky N, Braich R, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature 2005;438:685–689.
Nottrott S, Simard MJ, Richter JD . Human let-7a miRNA blocks protein production on actively translating polyribosomes. Nat Struct Mol Biol 2006;13:1108–1114.
Maroney PA, Yu Y, Fisher J, et al. Evidence that microRNAs are associated with translating messenger RNAs in human cells. Nat Struct Mol Biol 2006;13:1102–1107.
Pillai RS, Bhattacharyya SN, Artus CG, et al. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science 2005;309:1573–1576.
Wu L, Fan J, Belasco JG . MicroRNAs direct rapid deadenylation of mRNA. Proc Natl Acad Sci USA 2006;103:4034–4039.
Chendrimada TP, Finn KJ, Ji X, et al. MicroRNA silencing through RISC recruitment of eIF6. Nature 2007;447:823–828.
Standart N, Jackson RJ . MicroRNAs repress translation of m7Gppp-capped target mRNAs in vitro by inhibiting initiation and promoting deadenylation. Genes Dev 2007;21:1975–1982.
Vasudevan S, Tong Y, Steitz JA . Switching from repression to activation: microRNAs can upregulate translation. Science 2007;318:1931–1934.
Lee CT, Risom T, Strauss WM . Evolutionary conservation of MicroRNA regulatory circuits: an examination of MicroRNA gene complexity and conserved MicroRNA-target interactions through metazoan phylogeny. DNA Cell Biol 2007;26:209–218.
Griffiths-Jones S, Grocock RJ, van Dongen S, et al. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res 2006;34:D140–D144.
Clop A, Marcq F, Takeda H, et al. A mutation creating a potential illegitimate microRNA target site in the myostatin gene affects muscularity in sheep. Nat Genet 2006;38:813–818.
Walsh FS, Celeste AJ . Myostatin: a modulator of skeletal-muscle stem cells. Biochem Soc Trans 2005;33:1513–1517.
Mosher DS, Quignon P, Bustamante CD, et al. A mutation in the myostatin gene increases muscle mass and enhances racing performance in heterozygote dogs. PLoS Genet 2007;3:e79.
Sethupathy P, Borel C, Gagnebin M, et al. Human microRNA-155 on chromosome 21 differentially interacts with its polymorphic target in the AGTR1 3′ untranslated region: a mechanism for functional single-nucleotide polymorphisms related to phenotypes. Am J Hum Genet 2007;81:405–413.
Farh KK, Grimson A, Jan C, et al. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science 2005;310:1817–1821.
Voinnet O . Induction and suppression of RNA silencing: insights from viral infections. Nat Rev Genet 2005;6:206–220.
Landgraf P, Rusu M, Sheridan R, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 2007;129:1401–1414.
Poy MN, Eliasson L, Krutzfeldt J, et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature 2004;432:226–230.
Sempere LF, Freemantle S, Pitha-Rowe I, et al. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol 2004;5:R13.
Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature 2005;435:834–838.
Giraldez AJ, Cinalli RM, Glasner ME, et al. MicroRNAs regulate brain morphogenesis in zebrafish. Science 2005;308:833–838.
Houbaviy HB, Murray MF, Sharp PA . Embryonic stem cell-specific MicroRNAs. Dev Cell 2003;5:351–358.
Suh MR, Lee Y, Kim JY, et al. Human embryonic stem cells express a unique set of microRNAs. Dev Biol 2004;270:488–498.
Hatfield SD, Shcherbata HR, Fischer KA, et al. Stem cell division is regulated by the microRNA pathway. Nature 2005;435:974–978.
Forstemann K, Tomari Y, Du T, et al. Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol 2005;3:e236.
Murchison EP, Partridge JF, Tam OH, et al. Characterization of Dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci USA 2005;102:12135–12140.
Park JK, Liu X, Strauss TJ, et al. The miRNA pathway intrinsically controls self-renewal of drosophila germline stem cells. Curr Biol 2007;17:533–538.
Wang Y, Medvid R, Melton C, et al. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet 2007;39:380–385.
Murchison EP, Stein P, Xuan Z, et al. Critical roles for Dicer in the female germline. Genes Dev 2007;21:682–693.
Giraldez AJ, Mishima Y, Rihel J, et al. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science 2006;312:75–79.
Chen JF, Mandel EM, Thomson JM, et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet 2006;38:228–233.
Rao PK, Kumar RM, Farkhondeh M, et al. Myogenic factors that regulate expression of muscle-specific microRNAs. Proc Natl Acad Sci USA 2006;103:8721–8726.
Zhao Y, Ransom JF, Li A, et al. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell 2007;129:303–317.
Care A, Catalucci D, Felicetti F, et al. MicroRNA-133 controls cardiac hypertrophy. Nat Med 2007;13:613–618.
Naguibneva I, Ameyar-Zazoua M, Polesskaya A, et al. The microRNA miR-181 targets the homeobox protein Hox-A11 during mammalian myoblast differentiation. Nat Cell Biol 2006;8:278–284.
Chen CZ, Li L, Lodish HF, et al. MicroRNAs modulate hematopoietic lineage differentiation. Science 2004;303:83–86.
Li QJ, Chau J, Ebert PJ, et al. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell 2007;129:147–161.
Neilson JR, Zheng GX, Burge CB, et al. Dynamic regulation of miRNA expression in ordered stages of cellular development. Genes Dev 2007;21:578–589.
Thai TH, Calado DP, Casola S, et al. Regulation of the germinal center response by microRNA-155. Science 2007;316:604–608.
Rodriguez A, Vigorito E, Clare S, et al. Requirement of bic/microRNA-155 for normal immune function. Science 2007;316:608–611.
Vigorito E, Perks KL, Abreu-Goodger C, et al. MicroRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity 2007;27:847–859.
Zhan M, Miller CP, Papayannopoulou T, et al. MicroRNA expression dynamics during murine and human erythroid differentiation. Exp Hematol 2007;35:1015–1025.
Masaki S, Ohtsuka R, Abe Y, et al. Expression patterns of microRNAs 155 and 451 during normal human erythropoiesis. Biochem Biophys Res Commun 2007;364:509–514.
Rathjen T, Nicol C, McConkey G, et al. Analysis of short RNAs in the malaria parasite and its red blood cell host. FEBS Lett 2006;580:5185–5188.
Cao X, Yeo G, Muotri AR, et al. Noncoding RNAs in the mammalian central nervous system. Annu Rev Neurosci 2006;29:77–103.
Lagos-Quintana M, Rauhut R, Yalcin A, et al. Identification of tissue-specific microRNAs from mouse. Curr Biol 2002;12:735–739.
Li Y, Wang F, Lee JA, et al. MicroRNA-9a ensures the precise specification of sensory organ precursors in Drosophila. Genes Dev 2006;20:2793–2805.
Cao X, Pfaff SL, Gage FH . A functional study of miR-124 in the developing neural tube. Genes Dev 2007;21:531–536.
Visvanathan J, Lee S, Lee B, et al. The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev 2007;21:744–749.
Makeyev EV, Zhang J, Carrasco MA, et al. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell 2007;27:435–448.
Johnston RJ, Hobert O . A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature 2003;426:845–849.
Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA 2006;103:2257–2261.
Chan JA, Krichevsky AM, Kosik KS . MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res 2005;65:6029–6033.
Lui WO, Pourmand N, Patterson BK, et al. Patterns of known and novel small RNAs in human cervical cancer. Cancer Res 2007;67:6031–6043.
Ota A, Tagawa H, Karnan S, et al. Identification and characterization of a novel gene, C13orf25, as a target for 13q31–q32 amplification in malignant lymphoma. Cancer Res 2004;64:3087–3095.
He L, Thomson JM, Hemann MT, et al. A microRNA polycistron as a potential human oncogene. Nature 2005;435:828–833.
Dews M, Homayouni A, Yu D, et al. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat Genet 2006;38:1060–1065.
Karube Y, Tanaka H, Osada H, et al. Reduced expression of Dicer associated with poor prognosis in lung cancer patients. Cancer Sci 2005;96:111–115.
Kumar MS, Lu J, Mercer KL, et al. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet 2007;39:673–677.
Johnson SM, Grosshans H, Shingara J, et al. RAS is regulated by the let-7 microRNA family. Cell 2005;120:635–647.
Kluiver J, Haralambieva E, de Jong D, et al. Lack of BIC and microRNA miR-155 expression in primary cases of Burkitt lymphoma. Genes Chromosomes Cancer 2006;45:147–153.
Metzler M, Wilda M, Busch K, et al. High expression of precursor microRNA-155/BIC RNA in children with Burkitt lymphoma. Genes Chromosomes Cancer 2004;39:167–169.
Calin GA, Dumitru CD, Shimizu M, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA 2002;99:15524–15529.
Calin GA, Ferracin M, Cimmino A, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med 2005;353:1793–1801.
Cimmino A, Calin GA, Fabbri M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA 2005;102:13944–13949.
Fulci V, Chiaretti S, Goldoni M, et al. Quantitative technologies establish a novel microRNA profile of chronic lymphocytic leukemia. Blood 2007;109:4944–4951.
Tarasov V, Jung P, Verdoodt B, et al. Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR-34a is a p53 target that induces apoptosis and G(1)-arrest. Cell Cycle 2007;6:1586–1593.
Raver-Shapira N, Marciano E, Meiri E, et al. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell 2007;26:731–743.
He L, He X, Lim LP, et al. A microRNA component of the p53 tumour suppressor network. Nature 2007;447:1130–1134.
Chang TC, Wentzel EA, Kent OA, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell 2007;26:745–752.
Ma L, Teruya-Feldstein J, Weinberg RA . Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature 2007;449:682–688.
Clark EA, Golub TR, Lander ES, et al. Genomic analysis of metastasis reveals an essential role for RhoC. Nature 2000;406:532–535.
O'Hara A, Vahrson W, Dittmer DP . Gene alteration, pre- and mature micro RNA changes contribute to the miRNA signature of Primary Effusion Lymphoma (PEL). Blood 2008;111:2347–2353.
Gillis AJ, Stoop HJ, Hersmus R, et al. High-throughput microRNAome analysis in human germ cell tumours. J Pathol 2007;213:319–328.
Ozen M, Creighton CJ, Ozdemir M, et al. Widespread deregulation of microRNA expression in human prostate cancer. Oncogene 2008;27:1788–1793.
Roldo C, Missiaglia E, Hagan JP, et al. MicroRNA expression abnormalities in pancreatic endocrine and acinar tumors are associated with distinctive pathologic features and clinical behavior. J Clin Oncol 2006;24:4677–4684.
Blenkiron C, Goldstein LD, Thorne NP, et al. MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Biol 2007;8:R214.
Navarro A, Gaya A, Martinez A, et al. MicroRNA expression profiling in classical Hodgkin lymphoma. Blood 2008;111:2825–2832.
van Rooij E, Sutherland LB, Qi X, et al. Control of stress-dependent cardiac growth and gene expression by a MicroRNA. Science 2007;316:575–579.
van Rooij E, Sutherland LB, Liu N, et al. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci USA 2006;103:18255–18260.
Cai X, Schafer A, Lu S, et al. Epstein–Barr virus microRNAs are evolutionarily conserved and differentially expressed. PLoS Pathog 2006;2:e23.
Pfeffer S, Sewer A, Lagos-Quintana M, et al. Identification of microRNAs of the herpesvirus family. Nat Methods 2005;2:269–276.
Pfeffer S, Zavolan M, Grasser FA, et al. Identification of virus-encoded microRNAs. Science 2004;304:734–736.
Samols MA, Hu J, Skalsky RL, et al. Cloning and identification of a microRNA cluster within the latency-associated region of Kaposi's sarcoma-associated herpesvirus. J Virol 2005;79:9301–9305.
Cai X, Lu S, Zhang Z, et al. Kaposi's sarcoma-associated herpesvirus expresses an array of viral microRNAs in latently infected cells. Proc Natl Acad Sci USA 2005;102:5570–5575.
Sullivan CS, Grundhoff AT, Tevethia S, et al. SV40-encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cells. Nature 2005;435:682–686.
Skalsky RL, Samols MA, Plaisance KB, et al. Kaposi's sarcoma-associated herpesvirus encodes an ortholog of miR-155. J Virol 2007;81:12836–12845.
Stern-Ginossar N, Elefant N, Zimmermann A, et al. Host immune system gene targeting by a viral miRNA. Science 2007;317:376–381.
Jopling CL, Yi M, Lancaster AM, et al. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science 2005;309:1577–1581.
Lecellier CH, Dunoyer P, Arar K, et al. A cellular microRNA mediates antiviral defense in human cells. Science 2005;308:557–560.
Lu S, Cullen BR . Adenovirus VA1 noncoding RNA can inhibit small interfering RNA and MicroRNA biogenesis. J Virol 2004;78:12868–12876.
Haasnoot J, de Vries W, Geutjes EJ, et al. The Ebola virus VP35 protein is a suppressor of RNA silencing. PLoS Pathog 2007;3:e86.
Bennasser Y, Le SY, Benkirane M, et al. Evidence that HIV-1 encodes an siRNA and a suppressor of RNA silencing. Immunity 2005;22:607–619.
Li WX, Li H, Lu R, et al. Interferon antagonist proteins of influenza and vaccinia viruses are suppressors of RNA silencing. Proc Natl Acad Sci USA 2004;101:1350–1355.
O'Connell RM, Taganov KD, Boldin MP, et al. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci USA 2007;104:1604–1609.
Otsuka M, Jing Q, Georgel P, et al. Hypersusceptibility to vesicular stomatitis virus infection in Dicer1-deficient mice is due to impaired miR24 and miR93 expression. Immunity 2007;27:123–134.
Huang J, Wang F, Argyris E, et al. Cellular microRNAs contribute to HIV-1 latency in resting primary CD4+ T lymphocytes. Nat Med 2007;13:1241–1247.
Hafner M, Landgraf P, Ludwig J, et al. Identification of microRNAs and other small regulatory RNAs using cDNA library sequencing. Methods 2008;44:3–12.
Castoldi M, Benes V, Hentze MW, et al. MiChip: a microarray platform for expression profiling of microRNAs based on locked nucleic acid (LNA) oligonucleotide capture probes. Methods 2007;43:146–152.
Varallyay E, Burgyan J, Havelda Z . Detection of microRNAs by Northern blot analyses using LNA probes. Methods 2007;43:140–145.
Takada S, Mano H . Profiling of microRNA expression by mRAP. Nat Protoc 2007;2:3136–3145.
Lu C, Meyers BC, Green PJ . Construction of small RNA cDNA libraries for deep sequencing. Methods 2007;43:110–117.
Liu CG, Calin GA, Meloon B, et al. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proc Natl Acad Sci USA 2004;101:9740–9744.
Lim LP, Linsley PS . Mustering the micromanagers. Nat Biotechnol 2007;25:996–997.
Thompson RC, Deo M, Turner DL . Analysis of microRNA expression by in situ hybridization with RNA oligonucleotide probes. Methods 2007;43:153–161.
Nuovo GJ . In situ detection of precursor and mature microRNAs in paraffin-embedded, formalin-fixed tissues and cell preparations. Methods 2008;44:39–46.
Silahtaroglu AN, Nolting D, Dyrskjot L, et al. Detection of microRNAs in frozen tissue sections by fluorescence in situ hybridization using locked nucleic acid probes and tyramide signal amplification. Nat Protoc 2007;2:2520–2528.
Li J, Smyth P, Flavin R, et al. Comparison of miRNA expression patterns using total RNA extracted from matched samples of formalin-fixed paraffin-embedded (FFPE) cells and snap frozen cells. BMC Biotechnol 2007;7:36.
Xi Y, Nakajima G, Gavin E, et al. Systematic analysis of microRNA expression of RNA extracted from fresh frozen and formalin-fixed paraffin-embedded samples. RNA 2007;13:1668–1674.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Boyd, S. Everything you wanted to know about small RNA but were afraid to ask. Lab Invest 88, 569–578 (2008). https://doi.org/10.1038/labinvest.2008.32
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/labinvest.2008.32
Keywords
This article is cited by
-
Therapeutic angiogenesis with exosomal microRNAs: an effectual approach for the treatment of myocardial ischemia
Heart Failure Reviews (2021)
-
Integrative meta-analysis identifies microRNA-regulated networks in infantile hemangioma
BMC Medical Genetics (2016)
-
Transition from inflammation to proliferation: a critical step during wound healing
Cellular and Molecular Life Sciences (2016)
-
Reaction of small heat-shock proteins to different kinds of cellular stress in cultured rat hippocampal neurons
Cell Stress and Chaperones (2014)
-
LNA-based Oligonucleotide Electrotransfer for miRNA Inhibition
Molecular Therapy (2012)