Abstract
Kawasaki disease (KD), a systemic vasculitis of infants and children, manifests as fever and mucocutaneous inflammation. Although its etiology is largely unknown, the epidemiological data suggest that genetic factors are important in KD susceptibility. To identify genetic variants influencing KD susceptibility, we performed a genome-wide association study (GWAS) and replication study using a total of 915 children with KD and 4553 controls in the Korean population. Six single-nucleotide polymorphisms (SNPs) in three loci were associated significantly with KD susceptibility (P<1.0 × 10−5), including the previously reported BLK locus (rs6993775, odds ratio (OR)=1.52, P=2.52 × 10−11). The other two loci were newly identified: NMNAT2 on chromosome 1q25.3 (rs2078087, OR=1.33, P=1.15 × 10−6) and the human leukocyte antigen (HLA) region on chromosome 6p21.3 (HLA-C, HLA-B, MICA and HCP5) (rs9380242, rs9378199, rs9266669 and rs6938467; OR=1.33–1.51, P=8.93 × 10−6 to 5.24 × 10−8). Additionally, SNP rs17280682 in NLRP14 was associated significantly with KD with a family history (18 cases vs 4553 controls, OR=6.76, P=5.46 × 10−6). These results provide new insights into the pathogenesis and pathophysiology of KD.
Similar content being viewed by others
Introduction
Kawasaki disease (KD) is an acute, systemic vasculitis that affects mainly children under the age of 5. The clinical symptoms include the prolonged fever at least for more than 5 days and the following 5 clinical signs: bilateral conjunctival injection; erythema of the oral mucosa, lips, and tongue; polymorphous rash; erythema of the palms and soles; and cervical lymphadenopathy.1 It is well known that ~15–25% of untreated and 3–5% of treated children develop coronary artery aneurysms,2, 3, 4 making this disease the leading cause of acquired heart disease in infants and young children.
It is thought that KD develops in genetically susceptible children after being exposed by various immunological triggers, including an infectious agent(s).5, 6 More than 11–15 times higher incidence of KD is observed in Japan and Korea than in Caucasian population.7, 8, 9 Compared to children in Japan, in addition, almost same rate of KD incidence was also detected in Japanese–American residents in Hawaii.10 Furthermore, higher risk of KD occurrence was also observed in sibling cases and cases with parental history of KD.11, 12, 13 Ethnic difference and familial aggregation in KD incidence support that host genetic factors can contribute to disease susceptibility and outcome. It has been reported that ITPKC and CASP3 were identified as KD susceptibility genes by genome-wide linkage study and association studies.14, 15 In addition, genome-wide association studies (GWASs) reported four KD susceptibility genes: FCGR2A, BLK, CD40 and the HLA region, with genome-wide significance.16, 17, 18 These loci have provided a better understanding of the pathophysiology of KD. However, they do not fully explain the genetic risk of KD and some of them do not replicate in the Korean population. Therefore, we performed a GWAS to identify additional genetic risk factors for KD in the Korean population.
Materials and methods
Subjects
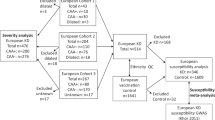
Children with KD were recruited from 12 tertiary academic hospitals in Korea that participated in the Korean Kawasaki Disease Genetics Consortium (KKDGC). Pediatricians diagnosed all KD patients according to the diagnostic criteria of the American Heart Association.19, 20 Control subjects were obtained from the adult healthy cohorts of the general population in Korea. These control samples were provided by the Biobank for Health Sciences at the Center for Genome Science in Chungwon, Korea. The GWAS was performed initially on 302 children with KD and 1000 controls. Among the cases, 253 had complete KD with a fever lasting 5 days or longer and more than four principal symptoms of KD, and 14 had a family history of KD. The replication study was performed on 720 KD cases, including 666 complete KD patients and four patients with a family history, and another 3553 controls. The second replication cohort from Japan comprised 428 KD cases and 3379 controls.17 The Institutional Review Boards of the involved institutions approved the study protocol, and all the parents of the KD patients provided written informed consent.
Genotyping and quality control
Genomic DNA was extracted from whole blood or lymphoblastoid cell lines according to standard protocols. For the GWAS, subjects were genotyped using the Illumina HumanOmni1-Quad BeadChip following the manufacturer’s instructions (Illumina, San Diego, CA, USA). All samples had a genotyping call rate of >98%. Six cases were excluded because of inconsistency between their gender on the clinical data sheets and that expected from the genotype data. All samples were unrelated to each other, based on an identity-by-state (IBS) analysis. A total of 296 KD cases and 1000 controls were included in subsequent analyses. To filter the single-nucleotide polymorphism (SNP) markers, we also excluded 488 SNP markers with missing call rates >2%, 43 markers with a Hardy–Weinberg Equilibrium (HWE) P-value <1 × 10−6 in the controls, and 208 625 markers with a minor allele frequency (MAF) <0.01. After filtering, 721,635 SNPs were included in the GWAS analysis. Genotyping for the replication study in the cases was performed on the high-throughput Fluidigm EP1 system (Fluidigm Corp., South San Francisco, CA, USA), using the Fluidigm SNPType assay platform and nanofluidic 96.96 and 48.48 Dynamic Array IFCs (Integrated Fluidic Circuits, Fluidigm). Genotype callings were made using the Fluidigm SNP Genotyping Analysis program (Fluidigm). We were able to genotype all of the selected SNPs for replication. The overall genotype success rate was 99.96%. Genotyping of the 3553 controls was conducted using the Illumina HumanOmni1-Quad BeadChip (Illumina). The Japanese population genotype data was imputed from the genotype data of the Illumina Human Hap550v3 BeadChip (Illumina).17
Statistical analysis
All statistical analyses were performed using the PLINK software (version 1.07) (http://pngu.mgh.harvard.edu/~purcell/plink/).21 A quantile–quantile plot and genomic inflation factor were used to assess the P-value distribution and population substructure. χ2 tests were used to compare allele frequencies between the cases and controls for genetic association study. Conditional analysis was carried out using logistic regression analysis under additive model. LD analysis (D′ and r2) and meta-analysis were also conducted with PLINK 1.07. Haploview 4.2 was used for drawing a Manhattan plot (https://www.broadinstitute.org/haploview/haploview). In addition, HaploReg v4.1 database (http://archive.broadinstitute.org/mammals/haploreg/haploreg.php) was used to search the quantitative trait loci (QTL) information of the KD-associated SNP rs2078087 in NMNAT2 locus.
Results
The NMNAT2 and HCP5 loci are associated with KD
To identify the genetic loci associated with KD susceptibility, we performed a GWAS using 249 children with KD and 1000 controls (Supplementary Table 1). The genomic control method showed no inflation of P-values (λ=1.047; Supplementary Figure 1). The GWAS results are shown in Supplementary Figure 2. Sixteen SNPs were associated with KD at P<1 × 10−5. For replication, we selected 98 representative SNPs considering their P-values (P<0.001 in at least two SNPs in one locus) and their immune-related functions and genotyped them using a Fluidigm SNPType assay platform. Among the 98 SNPs, 14 showed nominal evidence of replication (P<0.05) in 666 children with KD and 3553 controls (Supplementary Table 2). In the joint analysis of both the GWAS and the replication samples for these 14 SNPs, six SNPs in three loci were associated significantly with KD susceptibility at P<1 × 10−5 (Table 1). SNP rs6993775 with genome-wide significance (odds ratio (OR)=1.52, 95% confidence interval (95% CI)=1.33–1.72, P=2.52 × 10−11) was located in the intron of the BLK gene, which had been identified previously as a KD susceptibility locus from two recent GWASs.17, 18 The remaining five SNPs were identified for the first time in this study. SNP rs278087 (OR=1.33, 95% CI=1.19–1.50, P=1.15 × 10−6) was located in the intron of the NMNAT2 gene on chromosome 1q25.3 (Figure 1a), and the other four SNPs (rs6938467, rs9380242, rs9266669 and rs9378179) were mapped to the HLA class I region on chromosome 6p21.3 (Table 1, Figure 1b; P=8.93 × 10−6 to 5.24 × 10−8). Although each of the SNPs in the HLA region was located in a different gene (rs9380242 in HLA-C, rs9378199 in HLA-B, rs9266669 in MICA and rs6938467 in HCP5), when we analyzed the pair-wise linkage disequilibrium (LD), they all were linked to each other (D’=0.86–0.98, r2=0.42–0.91). After subsequent analysis conditioned on the most significant SNP in HCP5 (rs6938467), the significant associations at the other sites disappeared (Table 1), indicating that all four SNPs that were associated significantly with KD are located on the same LD block. We also performed a replication study in the Japanese population using the imputed genotype data of the Illumina Human Hap550v3 BeadChip in 428 KD cases and 3379 controls. SNP rs278087 in NMNAT2 showed a statistical trend (P=0.0608) and meta-analysis of Korean and Japanese populations showed a more significant association result (OR=1.27, 95% CI=1.16–1.40, P=5.90 × 10−7). However, the other SNPs in the HLA locus were not replicated in the Japanese population (Table 2).
Association plots of NMNAT2 and HLA genes. Shown are a regional association plot, the recombination rate and the linkage disequilibrium (LD) for the NMNAT2 region on chromosome 1q25.3 (a) and the HLA region on chromosome 6p21.3 (b), with gene annotations superimposed. Each single-nucleotide polymorphism (SNP) was plotted with respect to its chromosomal position (x axis) and its –log10 P-values (left, y axis) for the allelic test from the primary genome-wide association study (GWAS) scan (small circle). The significance levels in the combined analysis are also shown (large circle). The estimated recombination rates (right, y axis) based on the combined JPT and CHB samples from the 1000 genome project are plotted as a blue line. The color of each SNP symbol represents its LD (using the r2 algorithm) with the top SNP (large purple diamond) within the association locus. The image above was generated using the LocusZoom program (http://locuszoom.org/).
The NLRP14 locus is associated with KD with family history
Of the 296 children with KD, 14 had a family history of KD. Cases with a family history were more likely to be affected by genetic factors; therefore, we further investigated genetic loci affecting KD with a family history using 14 KD cases with a family history and 1000 controls. Six SNPs were chosen for replication according to the above criteria and tested in the remaining four cases and 3553 controls. In the combined analysis, rs17280682 at the NLRP14 locus was associated significantly with KD with a family history (OR=6.76, 95% CI=2.60–17.6, P=5.46 × 10−6; Table 3). However, the same SNP showed weak significance for the KD susceptibility in KD cases without a family history (993 cases vs 4553 controls) (P=0.0392). These results suggested that the NLRP14 locus affects the susceptibility of KD predominantly in genetically enriched familial cases.
Discussion
Using a large number of cases (n=915) and controls (n=4553) in a Korean population, we identified three known KD loci and two new KD-associated loci, rs2078087 in the NMNAT2 gene on chromosome 1q25.3, and rs6938467 in HLA class I region on chromosome 6p21.3, including the HLA-C, HLA-B, MICA and HCP5 genes (Table 1). We also identified the NLRP14 locus (rs17280682) on the chromosome 11p15.4 as being associated with KD with a family history (Table 3).
NMNAT2 is a member of the nicotinamide mononucleotide adenylyltransferase (NMNAT) enzyme family, which catalyze an essential step in the NAD biosynthetic pathway. A genetic variant (rs2022013) in NMNAT2 has been associated with susceptibility to systemic lupus erythematosus (SLE) (OR=0.85, P=1.08 × 10−7).22 Recently, Deng et al.23 confirmed the association of NMNAT2 with SLE, and identified an independent SLE signal at SMG7, adjacent to NMNAT2, tagged by rs2702178 (OR=1.23, P=2.4 × 10−8). SMG7 encodes a protein that is essential for nonsense-mediated mRNA decay. It has been also reported that a regulatory SNP (rs2275675) in the promoter region of SMG7 was associated robustly with SMG7 mRNA expression levels and its risk allele was significantly associated dose-dependently with decreased SMG7 mRNA levels in peripheral blood mononuclear cells (PBMCs) of patients with SLE and healthy controls, suggesting that differential SMG7 mRNA expression levels contribute to the SLE pathogenesis.23 SNP rs2022013 in NMNAT2 showed weak LD with rs2275675 in SMG7 (D′=0.61, r2=0.21); therefore, it is likely that they have independent effects on SLE susceptibility. In our combined analysis of the GWAS and replication samples, rs12144253 in the intron of SMG7 showed a weak but significant association with KD (OR=1.25, 95% CI=1.11–1.41, P=0.0003726). Therefore, both NMNAT2 and SMG7 are likely to have an effect on the susceptibility of KD. In this study, although we identified NMNAT2 locus as KD susceptibility, it is not clear why this enzyme in the NAD biosynthetic pathway is associated with KD and SLE. However, BLK locus showing the most significant association with KD was also reported as susceptibility gene for SLE,24, 25, 26 and rheumatoid arthritis.27 The association of BLK and NMNAT2 genes with both KD and SLE suggests that KD is sharing susceptibility genes with autoimmune diseases such as SLE and rheumatoid arthritis. In addition, the KD-associated SNP rs2078087 in NMNAT2 locus is associated with QTL for human metabolic trait, serum ratio of (1-oleoylglycerophosphocholine)/(16-anhydroglucose) (P=1.40 × 10−5; accessed by HaploReg), suggesting that the same SNP may also be involved in differential expression of NMNAT2 gene.
Four SNPs (rs9380242, rs9378199, rs9266669 and rs6938467) in the HLA class I region were associated with KD susceptibility (Table 1). Although they were located in different genes, they all were linked to each other (D′=0.86–0.98, r2=0.42–0.91). Furthermore, when we performed logistic regression analysis conditioned on the most significant SNP, rs6938467 in HCP5 (P=5.24 × 10−8), the significances at the other SNPs disappeared (Table 1), indicating that the associations at the four SNPs were not independent of each other. HCP5 encodes an endogenous retroviral element that has been reported to be associated with psoriasis,28 ulcerative colitis,29 HIV-1 control,30 AIDS progression,31 drug-induced liver injury due to flucloxacillin,32 Stevens–Johnson syndrome and toxic epidermal necrolysis (SJS-TEN)33, 34 and hypothyroidism.35 Variants in HCP5 have not been reported to be associated with KD susceptibility, whereas, variants in HLA-C, HLA-B and MICA were shown to be associated with KD susceptibility in candidate gene association studies of KD.36, 37, 38, 39, 40 All these genes are biologically critical for immunity and host defense, suggesting that all four genes are good candidates which can affect the pathogenesis of KD. However, this region contains highly polymorphic and complex genetic structure. Therefore, it is very difficult to reveal and discriminate a true signal from several linked association signals.
Furthermore, we showed that rs17280682 in NLRP14 was associated significantly with KD with a family history (Table 3). The protein encoded by NLRP14 gene belongs to the NALP (NACHT, LRR and PYD domains-containing protein 2) protein family and little is known about the function of NLRP14. However, it has been suggested that members of the NALP protein family play a role in apoptosis via activation of caspases and in proinflammation signaling processes.41, 42 In addition, the expression of NLRP3 inflammasome-associated genes, including NLRP1, NLRP3 and NLRP12 was upregulated during acute KD.43 Although the variants in the NLRP14 gene were not tested for their association with KD, it has been reported that one member of the NALP protein family, NLRP1, was associated with KD.44 This result supported the view that NLRP14 acts as a KD susceptibility gene, particularly in genetically enriched familial cases.
Recently, three GWASs reported five KD susceptibility loci exceeding the formal threshold for genome-wide significance: ITPKC, FCGR2A, BLK, HLA-DQB2 – HLA-DOB and CD40.16, 17, 18 We confirmed that the BLK locus was associated significantly with KD susceptibility in our Korean KD samples at a genome-wide significance level (Table 1; P=2.52 × 10−11), indicating that our genetic association study had sufficient power to detect genetic susceptibility loci in our sample sets. The FCGR2A and HLA-DOB loci also showed significant associations in our study (Table 1; P=5.65 × 10−5 in FCGR2A, 3.76 × 10−4 in HLA-DOB). However, the other two loci (ITPKC and CD40) were not replicated (data not shown). These observations suggested the presence of ethnic differences in susceptibility to KD. On the other hand, we also tested our findings in the Japanese population using the imputed genotype data of the Illumina Human Hap550v3 BeadChip in 428 KD cases and 3379 controls.17 rs278087 in NMNAT2 showed statistical trend (OR=1.17, P=0.0608) and meta-analysis of Korean and Japanese population showed more significant association result (OR=1.27, P=5.90 × 10−7). However, rs6938467 in HCP5 locus did not show significant association with KD (OR=1.15, P=0.1600) (Table 2). Therefore, further studies are necessary to validate our findings in other population using the real genotyping data with larger sample sets rather than the imputed genotype data.
In summary, we identified three known KD loci and two new KD-susceptibility loci in NMNAT2 and the HLA region in a Korean population. In addition, we identified the NLRP14 gene as a novel KD susceptibility locus involved in genetically enriched familial KD cases.
References
Burns, J. C. & Glodé, M. P. Kawasaki syndrome. Lancet 364, 533–554 (2004).
Kato, H., Sugimura, T., Akagi, T., Sato, N., Hashino, K., Maeno, Y. et al. Long-term consequences of Kawasaki disease. A 10- to 21-year follow-up study of 594 patients. Circulation 94, 1379–1385 (1996).
Newburger, J. W., Takahashi, M., Burns, J. C., Beiser, A. S., Chung, K. J., Duffy, C. E. et al. The treatment of Kawasaki syndrome with intravenous gamma globulin. N. Engl. J. Med. 315, 341–347 (1986).
Durongpisitkul, K., Gururaj, V. J., Park, J. M. & Martin, C. F. The prevention of coronary artery aneurysm in Kawasaki disease: a meta-analysis on the efficacy of aspirin and immunoglobulin treatment. Pediatrics 96, 1057–1061 (1995).
Newburger, J. W. & Fulton, D. R. Kawasaki disease. Curr. Opin. Pediatr. 16, 508–514 (2004).
Lee, K. Y., Han, J. W. & Lee, J. S. Kawasaki disease may be a hyperimmune reaction of genetically-susceptible children to variants of normal environmental flora. Med. Hypotheses 69, 642–651 (2007).
Makino, N., Nakamura, Y., Yashiro, M., Ae, R., Tsuboi, S., Aoyama, Y. et al. Descriptive epidemiology of Kawasaki disease in Japan, 2011-2012: from the results of the 22nd nationwide survey. J. Epidemiol. 25, 239–245 (2015).
Kim, G. B., Park, S., Eun, L. Y., Han, J. W., Lee, S. Y., Yoon, K. L. et al. Epidemiology and clinical features of Kawasaki disease in South Korea, 2012-2014. Pediatr. Infect. Dis. J. 36, 482–485 (2017).
Okubo, Y., Nochioka, K., Sakakibara, H., Testa, M. & Sundel, R. P. National survey of pediatric hospitalizations due to Kawasaki disease and coronary artery aneurysms in the USA. Clin. Rheumatol. 36, 413–419 (2017).
Holman, R. C., Curns, A. T., Belay, E. D., Steiner, C. A., Effler, P. V., Yorita, K. L. et al. Kawasaki syndrome in Hawaii. Pediatr. Infect. Dis. J. 24, 429–433 (2005).
Fujita, Y., Nakamura, Y., Sakata, K., Hara, N., Kobayashi, M., Nagai, M. et al. Kawasaki disease in families. Pediatrics 84, 666–669 (1989).
Harada, F., Sada, M., Kamiya, T., Yanase, Y., Kawasaki, T. & Sasazuki, T. Genetic analysis of Kawasaki syndrome. Am. J. Hum. Genet. 39, 537–539 (1986).
Uehara, R., Yashiro, M., Nakamura, Y. & Yanagawa, H. Kawasaki disease in parents and children. Acta Paediatr. 92, 694–697 (2003).
Onouchi, Y., Gunji, T., Burns, J. C., Shimizu, C., Newburger, J. W., Yashiro, M. et al. ITPKC functional polymorphism associated with Kawasaki disease susceptibility and formation of coronary artery aneurysms. Nat. Genet. 40, 35–42 (2008).
Onouchi, Y., Ozaki, K., Burns, J. C., Shimizu, C., Hamada, H., Honda, T. et al. Common variants in CASP3 confer susceptibility to Kawasaki disease. Hum. Mol. Genet. 19, 2898–2906 (2010).
Khor, C. C., Davila, S., Breunis, W. B., Lee, Y. C., Shimizu, C., Wright, V. J. et al. Genome-wide association study identifies FCGR2A as a susceptibility locus for Kawasaki disease. Nat. Genet. 43, 1241–1246 (2011).
Onouchi, Y., Ozaki, K., Burns, J. C., Shimizu, C., Terai, M., Hamada, H. et al. A genome-wide association study identifies three new risk loci for Kawasaki disease. Nat. Genet. 44, 517–521 (2012).
Lee, Y. C., Kuo, H. C., Chang, J. S., Chang, L. Y., Huang, L. M., Chen, M. R. et al. Two new susceptibility loci for Kawasaki disease identified through genome-wide association analysis. Nat. Genet. 44, 522–525 (2012).
Newburger, J. W., Takahashi, M., Gerber, M. A., Gewitz, M. H., Tani, L. Y. & Burns, J. C. et al. Diagnosis, treatment, and long-term management of Kawasaki disease: a statement for health professionals from the Committee on Rheumatic Fever, Endocarditis and Kawasaki Disease, Council on Cardiovascular Disease in the Young American Heart Association. Circulation 110, 2747–2771 (2004).
McCrindle, B. W., Rowley, A. H., Newburger, J. W., Burns, J. C., Bolger, A. F., Gewitz, M. et al. Diagnosis, treatment, and long-term management of Kawasaki disease: a scientific statement for health professionals from the American Heart Association. Circulation 135, e927–e999 (2017).
Purcell, S., Neale, B., Todd-Brown, K., Thomas, L., Ferreira, M. A., Bender, D. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
International Consortium for Systemic Lupus Erythematosus Genetics (SLEGEN) International Consortium for Systemic Lupus Erythematosus Genetics (SLEGEN), Harley, J. B. International Consortium for Systemic Lupus Erythematosus Genetics (SLEGEN), Alarcón-Riquelme, M. E. International Consortium for Systemic Lupus Erythematosus Genetics (SLEGEN), Criswell, L. A. International Consortium for Systemic Lupus Erythematosus Genetics (SLEGEN), Jacob, C. O. International Consortium for Systemic Lupus Erythematosus Genetics (SLEGEN), Kimberly, R. P. et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat. Genet. 40, 204–210 (2008).
Deng, Y., Zhao, J., Sakurai, D., Sestak, A. L., Osadchiy, V., Langefeld, C. D. et al. Decreased SMG7 expression associates with lupus-risk variants and elevated antinuclear antibody production. Ann. Rheum. Dis. 75, 2007–2013 (2016).
Graham, R. R., Cotsapas, C., Davies, L., Hackett, R., Lessard, C. J., Leon, J. M. et al. Genetic variants near TNFAIP3 on 6q23 are associated with systemic lupus erythematosus. Nat. Genet. 40, 1059–1061 (2008).
Hom, G., Graham, R. R., Modrek, B., Taylor, K. E., Ortmann, W., Garnier, S. et al. Association of systemic lupus erythematosus with C8orf13-BLK and ITGAM-ITGAX. N. Engl. J. Med. 358, 900–909 (2008).
Han, J. W., Zheng, H. F., Cui, Y., Sun, L. D., Ye, D. Q., Hu, Z. et al. Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nat. Genet. 41, 1234–1237 (2009).
Okada, T., Wu, D., Trynka, G., Raj, T., Terao, C., Ikari, K. et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature 506, 376–381 (2014).
Liu, Y., Helms, C., Liao, W., Zaba, L. C., Duan, S., Gardner, J. et al. A genome-wide association study of psoriasis and psoriatic arthritis identifies new disease loci. PLoS Genet. 4, e1000041 (2008).
Asano, K., Matsushita, T., Umeno, J., Hosono, N., Takahashi, A., Kawaguchi, T. et al. A genome-wide association study identifies three new susceptibility loci for ulcerative colitis in the Japanese population. Nat. Genet. 41, 1325–1329 (2009).
Fellay, J., Ge, D., Shianna, K. V., Colombo, S., Ledergerber, B., Cirulli, E. T. et al. Common genetic variation and the control of HIV-1 in humans. PLoS Genet. 5, e1000791 (2009).
Limou, S., Le Clerc, S., Coulonges, C., Carpentier, W., Dina, C., Delaneau, O. et al. Genomewide association study of an AIDS-nonprogression cohort emphasizes the role played by HLA genes (ANRS Genomewide Association Study 02). J. Infect. Dis. 199, 419–426 (2009).
Daly, A. K., Donaldson, P. T., Bhatnagar, P., Shen, Y., Pe'er, I., Floratos, A. et al. HLA-B*5701 genotype is a major determinant of drug-induced liver injury due to flucloxacillin. Nat. Genet. 41, 816–819 (2009).
Génin, E., Schumacher, M., Roujeau, J. C., Naldi, L., Liss, Y., Kazma, R. et al. Genome-wide association study of Stevens-Johnson Syndrome and toxic epidermal necrolysis in Europe. Orphanet J. Rare Dis. 6, 52 (2011).
Tohkin, M., Kaniwa, N., Saito, Y., Sugiyama, E., Kurose, K., Nishikawa, J. et al. A whole-genome association study of major determinants for allopurinol-related Stevens-Johnson syndrome and toxic epidermal necrolysis in Japanese patients. Pharmacogenomics J. 13, 60–69 (2011).
Eriksson, N., Tung, J. Y., Kiefer, A. K., Hinds, D. A., Francke, U., Mountain, J. L. et al. Novel associations for hypothyroidism include known autoimmune risk loci. PLoS ONE 7, e34442 (2012).
Kato, S., Kimura, M., Tsuji, K., Kusakawa, S., Asai, T., Juji, T. et al. HLA antigens in Kawasaki disease. Pediatrics 61, 252–255 (1978).
Krensky, A. M., Berenberg, W., Shanley, K. & Yunis, E. J. HLA antigens in mucocutaneous lymph node syndrome in New England. Pediatrics 67, 741–743 (1981).
Oh, J. H., Han, J. W., Lee, S. J., Lee, K. Y., Suh, B. K., Koh, D. K. et al. Polymorphisms of human leukocyte antigen genes in Korean children with Kawasaki disease. Pediatr. Cardiol. 29, 402–408 (2008).
Shrestha, S., Wiener, H. W., Aissani, B., Shendre, A., Tang, J. & Portman, M. A. Imputation of class I and II HLA loci using high-density SNPs from ImmunoChip and their associations with Kawasaki disease in family-based study. Int. J. Immunogenet. 42, 140–146 (2015).
Huang, Y., Lee, Y. J., Chen, M. R., Hsu, C. H., Lin, S. P., Sung, T. C. et al. Polymorphism of transmembrane region of MICA gene and Kawasaki disease. Exp. Clin. Immunogenet. 17, 130–137 (2000).
Inohara, N. & Nuñez, G. NODs: intracellular proteins involved in inflammation and apoptosis. Nat. Rev. Immunol. 3, 371–382 (2003).
Tschopp, J., Martinon, F. & Burns, K. NALPs: a novel protein family involved in inflammation. Nat. Rev. Mol. Cell Biol. 4, 95–104 (2003).
Alphonse, M. P., Duong, T. T., Shumitzu, C., Hoang, T. L., McCrindle, B. W., Franco, A. et al. Inositol-triphosphate 3-kinase c mediates inflammasome activation and treatment response in Kawasaki disease. J. Immunol. 197, 3481–3489 (2016).
Onoyama, S., Ihara, K., Yamaguchi, Y., Ikeda, K., Yamaguchi, K., Yamamura, K. et al. Genetic susceptibility to Kawasaki disease: analysis of pattern recognition receptor genes. Hum. Immunol. 73, 654–660 (2012).
Acknowledgements
We thank all the patients with Kawasaki disease and their families for participating in this study. This work was supported by a grant from the Ministry of Health & Welfare of the Republic of Korea (HI15C1575) and a grant from the Korea Center for Disease Control and Prevention (2014-ER7402-00).
Author information
Authors and Affiliations
Consortia
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Korean Kawasaki Disease Genetics Consortium Jeong Jin Yu3, In-Sook Park3, Soo-Jong Hong3, Kwi-Joo Kim3, Jong-Keuk Lee1, Jae-Jung Kim1, Young Mi Hong12, Sejung Sohn12, Gi Young Jang11, Kee-Soo Ha11, Hyo-Kyoung Nam11, Jung-Hye Byeon11, Sin Weon Yun2, Myung-Ki Han8, Kyung-Yil Lee5, Ja-Young Hwang5, Jung-Woo Rhim5, Min Seob Song9, Hyoung Doo Lee10, Dong Soo Kim20, Kyung Lim Yoon4, Hong-Ryang Kil6, Gi Beom Kim7, Jae-Moo Lee21 and Jong-Duk Kim21 20Department of Pediatrics, Yonsei University College of Medicine, Severance Children’s Hospital, Seoul, Korea; and 21Seoul Clinical Laboratories, Seoul, Korea
Supplementary Information accompanies the paper on Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Kim, JJ., Yun, S., Yu, J. et al. A genome-wide association analysis identifies NMNAT2 and HCP5 as susceptibility loci for Kawasaki disease. J Hum Genet 62, 1023–1029 (2017). https://doi.org/10.1038/jhg.2017.87
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2017.87
This article is cited by
-
Whole-exome sequencing analysis identifies novel variants associated with Kawasaki disease susceptibility
Pediatric Rheumatology (2023)
-
Whole-Exome Sequencing for Identification of Potential Sex-Biased Variants in Kawasaki Disease Patients
Inflammation (2023)
-
Association of an IGHV3-66 gene variant with Kawasaki disease
Journal of Human Genetics (2021)
-
Molecular mechanisms of long non-coding RNAs in anaplastic thyroid cancer: a systematic review
Cancer Cell International (2020)
-
Validation of genome-wide associated variants for Kawasaki disease in a Taiwanese case–control sample
Scientific Reports (2020)