Abstract
Fukuyama congenital muscular dystrophy (FCMD), which is caused by mutations in the fukutin gene, is the second most common form of childhood muscular dystrophy in Japan. The founder haplotype is the most prevalent in the chromosomes of Japanese FCMD patients, and corresponds to an SVA retrotransposal insertion in the 3′-untranslated region of fukutin. Although other mutations have been reported, the mutation corresponding to the second most prevalent haplotype in Japanese FCMD patients remained unknown. Recently a deep-intronic point mutation c.647+2084G>T was identified in Korean patients with congenital muscular dystrophy. Here, we performed mutational analysis of 10 patients with the second most prevalent haplotype and found that all of them were compound-heterozygous for the SVA insertion and this c.647+2084G>T mutation. The fukutin mRNA of these patients contained a pseudoexon between exon 5 and exon 6, which was consistent with the previous Korean study. As expected, the mutated fukutin protein was smaller than the normal protein, reflecting the truncation of fukutin due to a premature stop codon. Immunostaining analysis showed a decrease in the signal for the glycosylated form of α-dystroglycan. These findings indicated that this mutation is the second most prevalent loss-of-function mutation in Japanese FCMD patients.
Similar content being viewed by others
Introduction
Abnormal glycosylation of α-dystroglycan (α-DG) is known to cause some types of muscular dystrophy and lissencephaly, which are collectively called the α-dystroglycanopathies.1 Several causative genes of the α-dystroglycanopathies have been identified to date. Fukuyama congenital muscular dystrophy (FCMD) is a member of this disease group and is one of the most common autosomal recessive disorders in Japan.2 Patients with FCMD typically present with generalized hypotonia and weakness in infancy, followed by marked muscle atrophy, joint contractures and psychomotor developmental delay in childhood. Upright ambulation, even with support, is attained only rarely. The brain malformations are micropolygyria, pachygyria and agyria. Intellectual, cognitive and communicative functions are moderately delayed. Ophthalmological findings such as peripheral abnormalities of the retina or abnormal eye movements are often observed. The clinical course is inexorably progressive, with an average age at death of 16 years. We identified fukutin as the causative gene for FCMD through positional cloning,3 found that the clinical manifestations vary greatly among patients depending on the type of mutation in the fukutin gene,4 and demonstrated the possibility of splicing modulation therapy by antisense oligonucleotides as a clinical treatment for FCMD.5 A founder haplotype on chromosome 9q31 is seen in most FCMD patients (138-192-147-183, in terms of sizes of the PCR products of the markers D9S2105-D9S2170-D9S2171-D9S2107),6 and corresponds to an SVA retrotransposal insertion in the 3′-untranslated region of fukutin,3 which causes abnormal splicing.5 Several other mutations in FCMD patients have been found and linked to their own haplotypes. However, the mutation corresponding to the second most prevalent FCMD haplotype (139-201-155-183), which was found in nine out of the 107 patients heterozygous for the founder haplotype, has not been identified to date.4
In a recent report, mutations in fukutin were found to be responsible for the majority of Korean cases of congenital muscular dystrophy (CMD) with defective glycosylation of α-DG, and the two major types of fukutin mutations were the SVA insertion and a novel deep-intronic mutation (c.647+2084G>T).7 The latter point mutation occurred in intron 5, creating a new strong splicing donor site, and hence caused additional splicing to form a 64-bp pseudoexon between exon 5 and exon 6, and resulted in a frameshift and a premature stop codon. Considering the close ethnic interactions between Japan and Korea throughout history, we suspected that the second most prevalent haplotype in Japanese FCMD patients might be associated with this mutation identified in Korean patients, as in the case of the SVA insertion mutation, which is also shared between these two populations. We hence conducted a mutation analysis in 10 Japanese FCMD patients with the second most prevalent haplotype, to test this hypothesis.
Materials and methods
Nine patients with the second most common haplotype (139-201-155-183 for D9S2105-D9S2170-D9S2171-D9S2107) that were studied previously4 were analyzed in the present study. Another patient with this haplotype whose muscle biopsy specimen was available was also included. Cultured lymphoblastoid cell lines derived from three of the former nine patients were also available. All 10 patients have been genotyped and found to be heterozygous for this haplotype and the founder haplotype 138-192-147-183, which is linked to the SVA retrotransposal insertion.4 Parents of the patients who were available and willing to participate, one patient carrying the SVA retrotransposal mutation homozygously, two normal controls, and a person suspected of having a connective tissue disease were also included as controls. Informed consent was obtained from all participants. This study has been approved by the Human Ethics Review Committees of Kobe University Graduate School of Medicine and Tokyo Women’s Medical University.
Genomic DNA was extracted from peripheral blood. PCR was carried out with primers FKTNINT5f1 (CATGTGCAAAAATTTATCTTTGGCTATCTC) and FUKUTINT5r1 (GGTCATTTTGAAAATATGGCTTGGTTCAG) flanking the c.647+2084G>T mutation, using Ex Taq polymerase (Takara Bio, Shiga, Japan). RNA from lymphoblastoid cell lines was extracted using RNeasy plus kit (Qiagen, Venlo, The Netherlands), and RNA from skeletal musle was extracted using TRIzol Reagent (Invitrogen, Carlsbad, California, US). RT-PCR with the primer pair FCMDex4F (CATGCGATCCACTTGGTAGTC) and FCMDex6-7 (GGTACTGCTGAAAGAATGCTCG) was carried out with SuperScript III Reverse Transcriptase (Invitrogen) and Ex Taq polymerase. PCR products were purified and direct-sequenced. PCR-restriction fragment length polymorphism (RFLP) analysis to detect the c.647+2084G>T mutation was performed as follows: a mismatched primer pair was designed so that the PCR product consistently contains an Rsa I restriction enzyme recognition site (GTAC) in the 5′ part as a positive control for Rsa I digestion, and also in the 3′ part only when the template DNA carries the c.647+2084G>T mutation, namely, primers FKTN5f3M (GGATTAAAAACATTCTTGAAGTTATACTTGGAGTActAAGTTTC) and FKTN5rM (CTCACTGGAAGTTACTAAGAAGGAGTTTTTAATGTGAAAgT) (the mismatched sequences are written in lowercase letters). Genomic DNA was amplified with the mismatched primer pair using AmpliTaq Gold master mix (Applied Biosystems, Foster City, CA, USA), and the PCR product, with a length of 195 bp, was then treated with Rsa I. The product was digested into two fragments of 32 and 163 bp, and if the c.647+2084G>T mutation is present, the 163-bp fragment is additionally digested to 120-bp and 43-bp fragments. Thus, we can conclude that a sample carries the c.647+2084G>T mutation if the 120-bp fragment is observed. The SVA retrotransposal insertion and the endogenous fukutin protein were detected as described previously.5, 8 Immunostaining of muscle biopsy cryosections was performed using antibodies against α-DG (IIH6: Millipore, Billerica, MA, USA; AP-074G-C, reported previously.9 Briefly, AP-074G-C was raised in Goat against recombinant α-DG lacking a signal sequence and the mucin domain that was produced in Escherichia coli, and affinity-purified using recombinant α-DG secreted from HEK293.), β-dystroglycan (β-DG) (8D5: Novocastra, Newcastle upon Tyne, UK), dystrophin (ab15277: Abcam, Cambridge, MA, USA), and laminin-α2 (4H8-2: Abcam).
Results
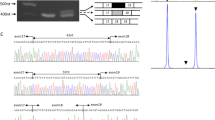
We first attempted direct sequencing of the PCR products from genomic DNA of three (patient no. 7, 8 and 9) of the 10 patients with the haplotype 139-201-155-183 for D9S2105-(FCMD)-D9S2170-D9S2171-D9S2107 around the fukutin gene, to check if these patients carried the deep-intronic point mutation c.647+2084G>T in fukutin that was previously found in Korean CMD patients.7 We found that all three patients carried this point mutation in heterozygous form (data not shown). We then constructed a PCR-RFLP system using a mismatched primer pair to detect c.647+2084G>T and applied this method to the 10 patients with the haplotype 139-201-155-183 and their parents, together with analysis of the SVA retrotransposal insertion (Figure 1). The results showed that all 10 patients have the c.647+2084G>T mutation and clearly demonstrated the parental origins of the c.647+2084G>T mutation and the SVA insertion in each patient. Considering their haplotypes that had been analyzed previously, we concluded that the haplotype 139-201-155-183 is linked to the c.647+2084G>T mutation in intron 5 of the fukutin gene.4
Mutation status of the DNA of patients carrying the second most common haplotype and the founder haplotype heterozygously. Upper panels show the results of PCR-RFLP analysis to detect the c.647+2084G>T mutation. The lower bands (120 bp) demonstrate the existence of the mutation. Lower panels show the results of PCR analysis to detect the SVA insertion mutation. The upper bands (375 bp) demonstrate the existence of the insertion. M: 50-bp ladder DNA marker; 1–10: patient numbers.
Next, we performed RT-PCR and direct sequencing using RNA from the cultured lymphoblastoid cell lines of patients no. 7, 8 and 9. We found that fukutin RNA of all three patients carried the 64-bp pseudoexon insertion between exon 5 and exon 6 that is caused by abnormal splicing, which is consistent with the previous report (Figure 2).7 We also confirmed that RNA from skeletal muscle of patient no. 10 also contains this pseudoexon.
RNA status of the patients carrying the c.647+2084G>T mutation. (a) Upper bands (347 bp) show the RT-PCR products containing a pseudoexon. (b) Schematic representation of the abnormal splicing caused by this mutation, which results in the pseudoexon. (c) The nucleotide sequences of the pseudoexon and the flanking intron parts. The capital letters represent the pseudoexon sequence and the small letters represent the flanking intron sequence. A premature stop codon is underlined. The c.647+2084G>T mutation is shown as a bold letter. M: 50-bp ladder DNA marker; 7–10: patient numbers; N: normal control; H: a patient carrying the SVA insertion homozygously.
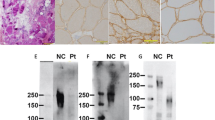
The pseudoexon insertion is thought to cause a frameshift and a premature stop codon. To confirm that this mutation results in the synthesis of a shorter fukutin protein (presumably ~26 kDa, compared with ~55 kDa for normal fukutin and ~62 kDa for mutant fukutin from the SVA insertion allele), we performed immunoprecipitation and western blotting to detect the endogenously expressed fukutin protein in cultured lymphoblastoid cell lines derived from patients no. 7, 8 and 9 (Figure 3). A larger-sized band derived from the heterozygous SVA insertion allele was observed in all patients as described previously,5 and, consistent with our prediction, another smaller-sized weak band (~26 kDa) was also observed exclusively in the three patients carrying the pseudoexon insertion. The expression level of the truncated fukutin seems to be very low probably due to the nonsense-mediated mRNA decay mechanism.
Fukutin protein status of patients carrying the c.647+2084G>T mutation. The upper panel shows the normal fukutin protein (~55 kDa) and the mutant fukutin protein (~62 kDa) derived from the SVA insertion allele. The lower panel shows the truncated fukutin protein that is possibly derived from the c.647+2084G>T mutation allele, detected by long exposure. H: a patient carrying the SVA insertion homozygously; N: normal control; 7–9: patient numbers.
We also performed immunostaining to assess the effects of the c.647+2084G>T mutation on the integrity of the dystrophin-glycoprotein complex in the skeletal muscle from one patient (no. 10). Components of the dystrophin-glycoprotein complex, including dystrophin, β-DG, α-DG and laminin-α2 were analyzed (Figure 4). All of these proteins showed normal distributions and expression; however, the glycosylated form of α-DG was not detected, indicating that c.647+2084G>T causes the abnormal glycosylation of α-DG. The α-DG core protein seemed to be slightly reduced in muscle of this patient, although the staining of the α-DG core protein seemed to be normal in previous report.10 The mechanism behind this discrepancy has yet to be clarified, and it might be due to the variability in the strength of staining depending on patient’s background or something. Hematoxylin and eosin staining showed dystrophic changes, including fiber size variability and fibrosis (data not shown).
Discussion
In our previous study, 80 out of 107 Japanese FCMD patients were homozygous for the founder haplotype (138-192-147-183), which is linked to a 3-kb SVA retrotransposal insertion mutation in the fukutin gene.4 Twenty-five of the remaining 27 patients were compound-heterozygous for the founder haplotype and another haplotype. Nine of the 25 patients carried the second most common haplotype 139-201-155-183; however, a specific mutation linked to this haplotype has not yet been found. In this study, we successfully showed that c.647+2084G>T in intron 5 of the fukutin gene is linked to this haplotype. Seven of the 25 patients had the third most common haplotype 130-201-157-183, which corresponds to the c.139C>T nonsense mutation in exon 3 of the fukutin gene. Therefore, these three mutations account for approximately 90% (96/107) of the mutations of Japanese FCMD patients. The PCR-RFLP method to detect the c.647+2084G>T mutation mentioned in this study will be clinically useful for genetic testing of FCMD patients, together with the PCR method to detect the SVA insertion and the PCR-RFLP method to detect the c.139C>T nonsense mutation.3 We previously demonstrated that splicing modulation therapy by antisense oligonucleotides is a possible radical treatment for FCMD.5 Patients who are compound-heterozygous for the SVA insertion and c.647+2084G>T are also expected to benefit from this potential therapy.
Patients carrying the c.647+2084G>T mutation clinically present with severe phenotypes, such as no head control or sitting with support, compared with patients carrying the SVA insertion homozygously.4 This might be explained by the sites of abnormal splicing. Both the c.647+2084G>T mutation and SVA insertion cause abnormal splicing. The normal fukutin gene consists of 10 exons. The c.647+2084G>T mutation results in a pseudoexon between exon 5 and exon 6 and produces a half-truncated fukutin protein that does not contain the DXD motif, which is a putative catalytic site of the fukutin protein (p.Arg216Serfs*10). On the other hand, a splicing acceptor site located within the SVA insertion in exon 10 activates a rare alternative splicing donor site in exon 10, causes abnormal splicing involving the authentic stop codon, and produces a truncated fukutin protein lacking a small part of the C-terminus but containing the DXD motif (p.Gly423_Tyr461delins129).5 Recently, we found that fukutin is a ribitol-phosphate transferase that is essential for laminin-binding glycan synthesis on α-DG.11 Fukutin transfers ribitol-phosphate from CDP-ribitol that is produced by ISPD, one of the gene products responsible for the α-dystroglycanopathies, to the C3 position of N-acetylglucosamine at the top of the phosphorylated O-mannose glycan called CoreM3 on α-DG. Fukutin-related protein, FKRP, another α-dystroglycanopathy gene product, sequentially transfers ribitol-phosphate from CDP-ribitol to the C1 position of ribitol-phosphate which is transferred by fukutin. Thus, the former mutant fukutin protein produced by the c.647+2084G>T mutation may be dysfunctional, whereas the latter by the SVA insertion may retain some enzyme activity. The difference in the small amount of normal fukutin protein that is produced from the very small amount of normally spliced products may be another possible reason.
FCMD was once thought to be endemic only in Japan, but many non-Japanese patients have been found to have mutations in the fukutin gene. Moreover, the SVA insertion, which is the Japanese founder mutation, has been found in several Korean and Chinese CMD patients.12, 13 Thus, it was not beyond our expectation that the c.647+2084G>T mutation is shared between Japanese and Korean patients, considering their ethnic interactions throughout history. When and where this mutation appeared and how it spread among the modern Japanese and Korean population might be of great interest from the view of ethnology. As for the SVA insertion, its age was calculated to be ~100 generations in our previous study, indicating that it emerged in the Yayoi period when immigrants from Asian continents settled in the Japanese islands.14 For such discussion regarding the c.647+2084G>T mutation, haplotype analysis of Korean patients will offer a large amount of useful information. If Korean patients with the c.647+2084G>T mutation have the same haplotype (i.e., 139-201-155-183), these patients probably originate from a common ancestor with Japanese patients carrying this mutation.
References
Yoshida-Moriguchi, T. & Campbell, K. P. Matriglycan: a novel polysaccharide that links dystroglycan to the basement membrane. Glycobiology 25, 702–713 (2015).
Fukuyama, Y. & Ohsawa, M. A genetic study of the Fukuyama type congenital muscular dystrophy. Brain Dev. 6, 373–390 (1984).
Kobayashi, K., Nakahori, Y., Miyake, M., Matsumura, K., Kondo-Iida, E., Nomura, Y. et al. An ancient retrotransposal insertion causes Fukuyama-type congenital muscular dystrophy. Nature 394, 388–392 (1998).
Kondo-Iida, E., Kobayashi, K., Watanabe, M., Sasaki, J., Kumagai, T., Koide, H. et al. Novel mutations and genotype-phenotype relationships in 107 families with Fukuyama-type congenital muscular dystrophy (FCMD). Hum. Mol. Genet. 8, 2303–2309 (1999).
Taniguchi-Ikeda, M., Kobayashi, K., Kanagawa, M., Yu, C. C., Mori, K., Oda, T. et al. Pathogenic exon-trapping by SVA retrotransposon and rescue in Fukuyama muscular dystrophy. Nature 478, 127–131 (2011).
Kobayashi, K., Nakahori, Y., Mizuno, K., Miyake, M., Kumagai, T., Honma, A. et al. Founder-haplotype analysis in Fukuyama-type congenital muscular dystrophy (FCMD). Hum. Genet. 103, 323–327 (1998).
Lim, B. C., Ki, C. S., Kim, J. W., Cho, A., Kim, M. J., Hwang, H. et al. Fukutin mutations in congenital muscular dystrophies with defective glycosylation of dystroglycan in Korea. Neuromuscul. Disord. 20, 524–530 (2010).
Watanabe, M., Kobayashi, K., Jin, F., Park, K. S., Yamada, T., Tokunaga, K. et al. Founder SVA retrotransposal insertion in Fukuyama-type congenital muscular dystrophy and its origin in Japanese and Northeast Asian populations. Am. J. Med. Genet. 138A, 344–348 (2005).
Kanagawa, M., Nishimoto, A., Chiyonobu, T., Takeda, S., Miyagoe-Suzuki, Y., Wang, F. et al. Residual laminin-binding activity and enhanced dystroglycan glycosylation by LARGE in novel model mice to dystroglycanopathy. Hum. Mol. Genet. 18, 621–631 (2009).
Michele, D. E., Barresi, R., Kanagawa, M., Saito, F., Cohn, R. D., Satz, J. S. et al. Post-translational disruption of dystroglycan–ligand interactions in congenital muscular dystrophies. Nature 418, 417–422 (2002).
Kanagawa, M., Kobayashi, K., Tajiri, M., Manya, H., Kuga, A., Yamaguchi, Y. et al. Identification of a post-translational modification with ribitol-phosphate and its defect in muscular dystrophy. Cell Rep. 14, 2209–2223 (2016).
Lee, J., Lee, B. L., Lee, M., Kim, J. H., Kim, J. W., Ki, C. S. et al. Clinical and genetic analysis of a Korean patient with Fukuyama congenital muscular dystrophy. J. Neurol. Sci. 281, 122–124 (2009).
Xiong, H., Wang, S., Kobayashi, K., Jiang, Y., Wang, J., Chang, X. et al. Fukutin gene retrotransposal insertion in a non-Japanese Fukuyama congenital muscular dystrophy (FCMD) patient. Am. J. Med. Genet. 149A, 2403–2408 (2009).
Colombo, R., Bignamini, A. A., Carobene, A., Sasaki, J., Tachikawa, M., Kobayashi, K. et al. Age and origin of the FCMD 3′-untranslated-region retrotransposal insertion mutation causing Fukuyama-type congenital muscular dystrophy in the Japanese population. Hum. Genet. 107, 559–567 (2000).
Acknowledgements
We thank Ms Hiroko Nakano for the PCR and PCR-RFLP experiments; and Dr Helena Akiko Popiel and Ms Kazumi Ura for editing the manuscript. This work was supported by grants from the National Center of Neurology and Psychiatry (Intramural Research Grant 26-8 to TT) and Japan Society for the Promotion of Science (grant no 26253057 to TT, and 26670499 and 16H05353 to KK), and by the Practical Research Project for Rare/Intractable Diseases (Step1) from Japan Agency for Medical Research and development, AMED (17930060 to TT).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Kobayashi, K., Kato, R., Kondo-Iida, E. et al. Deep-intronic variant of fukutin is the most prevalent point mutation of Fukuyama congenital muscular dystrophy in Japan. J Hum Genet 62, 945–948 (2017). https://doi.org/10.1038/jhg.2017.71
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2017.71
This article is cited by
-
International consensus recommendations on the diagnostic work-up for malformations of cortical development
Nature Reviews Neurology (2020)