Abstract
Wilson’s disease (WD) is an autosomal recessive disorder caused by ATP7B gene mutation. The frequency of WD is about 1 in 30 000 worldwide. In the present study, we screened 14 835 dried blood spots (DBSs) from asymptomatic Korean neonates and retrospectively reviewed massively parallel sequencing of 1090 control individuals to estimate carrier frequency. TaqMan real-time PCR assays were conducted to detect six mutations that account for 58.3% of mutations in Korean WD patients: c.2333G>T (p.Arg778Leu), c.2621C>T (p.Ala874Val), c.3086C>T (p.Thr1029Ile), c.3247C>T (p.Leu1083Phe), c.3556G>A (p.Gly1186Ser) and c.3809A>G (p.Asn1270Ser). We also retrospectively reviewed data from 1090 individuals with various indications other than WD for whom whole-exome or panel sequencing data were available. Mutant allele frequency based on the six most common mutations was 0.0067 among the total of 14 835 DBSs screened. Given that these six mutations account for 58.3% of mutations in Korean WD patients, the corrected mutant allele frequency is 0.0115 (95% confidence interval (CI): 0.0103–0.0128). Corresponding incidence (q2) and carrier frequency (2pq) were estimated to be 1:7561 and 1:44, respectively. In retrospective data analysis of 1090 control individuals, allele frequency of pathogenic or likely pathogenic variants was 0.0096 (95% CI: 0.0063–0.0146). Corresponding carrier frequency was estimated to be 1:53. Estimated allele and carrier frequencies based on DNA screening were relatively higher than those reported previously based on clinical ascertainment.
Similar content being viewed by others
Introduction
Wilson’s disease (WD) is an autosomal recessive disorder caused by mutation of the ATP7B gene, which encodes a protein called copper-transporting ATPase 2. The defect results in copper accumulation in the liver due to reduced excretion of copper into bile. WD can manifest as hepatic, neurologic or psychiatric symptoms. However, most patients do not have all of these findings. Manifestations are often nonspecific for clinical suspicion of WD. The presence of Kayser–Fleisher rings, caused by deposition of copper in the periphery of the cornea, highly suggests WD. However, this symptom is observed in only 50–60% of patients with liver disease, although ~90% of individuals with either neurologic or psychiatric disturbance have Kayser–Fleisher rings. Therefore, some WD patients might not be correctly diagnosed.
About 760 unique ATP7B mutations have been reported (Human Gene Mutation Database), with mutations dispersed across exons. Recurrent mutations are found in some populations; the H1069Q mutation is the most common allele in patients of European descent,1 whereas deletion of a 5′-untranslated region (−441/−427 del) is common in WD patients of Sardinian descent.2 The R778L mutation is the most prevalent in Koreans3, 4, 5 and Chinese,6 whereas 2871delC or 2874delC mutations are recurrent mutations in the Japanese population.7, 8 Some studies have estimated carrier frequency using recurrent mutations.9 Because WD is thought to be one of the most prevalent autosomal recessive diseases in Koreans, the carrier frequencies of certain ATP7B mutations in the Korean population have been studied previously using DNA-based approaches. However, these studies were based on a limited number of samples.10, 11, 12 In the present study, we developed a robust DNA-based screening for WD based on the TaqMan real-time PCR method that is applicable to dried blood spots (DBSs). Based on this method, we screened 14 835 DBSs from asymptomatic Korean neonates and estimated the carrier frequency of WD in this population. In addition, we retrospectively reviewed massively parallel sequencing (MPS) data from 1090 individuals to estimate carrier frequency based on whole exons of the ATP7B gene.
Materials and methods
Patients
Samples were referred from obstetric hospitals for WD screening from December 2014 to April 2016. Information was given to the parents regarding WD and the genetic screening test. The test was conducted at the parents’ initiative on a self-pay basis. For the genetic screening test, DBSs were obtained from the neonate. If the neonate was found to have one pathogenic variant among the tested loci, the parents were counseled about the residual risk of having pathogenic variants in untested regions of the ATP7B gene. The parents were given the further option of whole exon sequencing either by direct sequencing or MPS.
Mutation screening by TaqMan real-time PCR
Genomic DNA was extracted from the DBSs using the GeneAll DNA Extraction Kit (GeneAll Biotechnologies, Seoul, Korea) according to the manufacturer’s instructions. Three to five disks with a 1.5 mm diameter were manually punched out per sample. TaqMan real-time PCR assays were performed to detect six mutations: c.2333G>T (p.Arg778Leu), c.2621C>T (p.Ala874Val), c.3086C>T (p.Thr1029Ile), c.3247C>T (p.Leu1083Phe), c.3556G>A (p.Gly1186Ser) and c.3809A>G (p.Asn1270Ser). Selection of the mutations was based on previous knowledge about the prevalence of pathogenic variants in Korean WD patients.5, 10, 12 We estimated that analysis of these six mutations would cover 58.3% of pathogenic variants in the Korean population. Primers and probes were ready-made (Thermo Fisher Scientific, Waltham, MA, USA) or customized according to the literature. PCR was performed on a CFX96 (Bio-Rad, Hercules, CA, USA), and cycle conditions were as follows: an initial denaturation step of 95 °C for 1 min, followed by 40 cycles of denaturation at 95 °C for 15 s and primer annealing and extension at 60 °C for 30 s.
Retrospective data review
We retrospectively reviewed anonymized data from 1090 individuals with various indications other than WD for whom whole-exome or panel sequencing had been conducted. Whole-exome sequencing data from 300 individuals and panel sequencing data from 790 individuals were included. Whole-exome sequencing was performed with SureSelect (Agilent Technologies, Santa Clara, CA, USA), and panel sequencing was performed with TruSight One Sequencing Panel (Illumina, San Diego, CA, USA), which captures 4813 genes with oligonucleotide probes. MPS was performed using 2 × 150 bp in the paired-end mode of the NextSeq platform (Illumina). Sequence reads were aligned with the Burrow-Wheeler Aligner (version 0.7.5). After duplicated reads were removed with Picard, local realignment and recalibration were performed with the Genome Analysis Tool Kit (GATK, version 1.6). Variant calling was conducted with GATK, and variants were annotated with the human mutation database, SNP databases (dbSNP, 1000 Genomes Project, ESP 6500, ExAC) and Korean Reference Genome Database, which is a whole-genome sequencing data set from 622 Korean individuals released by the Korean National Research Institute of Health. Among the variants, minor alleles with a frequency exceeding 1% were excluded. For variants to be classified as pathogenic, they had to be classified as disease-causing mutations in the Human Gene Mutation Database or be truncating or splice site mutations. Novel missense variants were classified based on in silico analysis (SIFT, PolyPhen-2) and the conservation score (GERP). If the predictions of the two missense prediction tools were consistent and the DNA sequence was conserved (GERP++ RS >4.4),13 the variant was classified as likely pathogenic. Variants with a minimum depth of 25 × with a frequency of heterozygous variants ranging from 0.25 to 0.75 were included, and visual inspection of variants was performed. The study was conducted in accordance with the Declaration of Helsinki of 1975, as revised in 2013.
Statistical analysis
For the calculation of 95% confidence interval (CI) of the mutant allele frequency, the online software was used (http://www.vassarstats.net/prop1.html).
Results
Allele frequency of six mutations based on TaqMan real-time PCR
A total of 14 835 DBSs were analyzed for six mutations. The most common mutation was R778L, with an allele frequency of 0.0033. The allele frequency of all six mutations was 0.0067. Allele frequencies of individual mutations are shown in Table 1.
Because the six mutations account for 58.3% of mutations in Korean WD patients according to a previous study,5 the allele frequency of these six mutations is effectively 0.0115 (95% CI: 0.0103–0.0128). Corresponding incidence (q2) and carrier frequency (2pq) were estimated to be 1:7561 (0.00013225) and 1:44 (0.0227355), respectively.
Retrospective data set
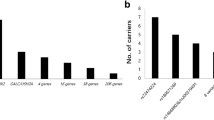
Among 1090 control individuals, a total of 87 variants were present in the ATP7B gene. After filtering based on a minor allele frequency of 1% using the public database and Korean-specific genome database, 27 unique variants remained. Among these, five of six mutations included in the TaqMan real-time PCR were detected in 11 individuals; seven individuals were heterozygous for p.R778L, whereas p.A874V, p.T1029I, p.L1083F and p.N1270S were detected only once each. p.G1186S was not detected in this data set. The allele frequency of these six mutations was 0.005. In addition, six other known mutations and one novel mutation were detected once in a heterozygous form (Table 2). Therefore, the allele frequency of definitely pathogenic variants was 0.0083 (18/2,180). The p.Val1106Ile variant, which is classified as a likely pathogenic variant in the Human Gene Mutation Database, was suggested to be a polymorphism by an in vitro functional study,5 and the p.Ala476Thr variant is classified as a likely benign variant in the ClinVar database (assessed on 26 December 2016). Among the remaining 12 unique variants, one (p.Glu316Lys) found in three individuals was predicted to be deleterious by in silico analysis. The mutant allele frequency increased to 0.0096 (95% CI: 0.0063–0.0146) when likely pathogenic variants supported by in silico analysis were included.
Discussion
The allele frequency of the six most common WD mutations in the Korean population was estimated to be 0.0067 based on screening of 14 835 DBSs; this result is similar to that of Song et al.12 (Table 1). Although previous studies reported allele frequencies of 0.0032 and 0.01 based on the analysis of three mutations (p.R778L, p.A874V and p.N1270S),10, 11 these data may be biased by the small number of subjects included in those previous studies. Considering that the six mutations screened for in DBSs account for 58.3% of mutations detected in Korean WD patients, we converted the mutant allele frequency and carrier frequency to 0.0115 and 1:44 (0.0227355), respectively. Therefore, the frequency of WD is 1:7561 (0.00013225). In our retrospective data set that included whole coding exons of the ATP7B gene, the allele frequency of pathogenic or likely pathogenic variants was 0.0096, and the corresponding carrier frequency was 1:53 (0.01901568). Our data are comparable to those of a recent study that performed whole coding exon sequence analysis of 1000 DNA samples. In that study, the carrier frequency was reported to be 1/42 (0.024), and the calculated frequency of individuals carrying two pathogenic variants was 1:7026.14 This is significantly different from the carrier frequency of 1/90 individuals calculated using Hardy–Weinburg equilibrium based on the known WD frequency of 1/30 000 individuals.15 This suggests that WD might be underdiagnosed.
Early detection of WD can benefit the patient because avoiding foods containing high levels of copper and taking copper-chelating agents can prevent irreversible damage. The biomarker currently used for screening is ceruloplasmin level, either in blood or urine; however, some symptomatic patients have normal ceruloplasmin level. For early detection of WD, screening tests targeting high-risk populations such as those with liver disease or mass screening have been attempted.16, 17, 18, 19 Other studies have screened for WD using DBSs in populations ranging from newborns to adolescents,16, 18 and one WD patient was detected among 3667 individuals screened. However, determination of ceruloplasmin by enzyme-linked immunosorbent assay has the limitations of low repeatability or reproducibility;18 only 10% of samples that had a low ceruloplasmin level in the first test were consistently abnormal in the second test.16 In addition, a low level of ceruloplasmin can be found in some WD carriers (~10%) and in diseases other than WD such as copper deficiency, Menkes disease and hereditary aceruloplasminemia. Ceruloplasmin level can also be elevated by inflammatory conditions as an acute-phase reactant. This has led to increased interest in DNA-based screening tests.20
Population screening based on recurrent mutations has been suggested. Whole exon sequencing should be performed to screen positive samples to determine the presence of second mutations to discriminate between heterozygous carriers and affected patients. Recently, MPS has been used for diagnosis and/or screening purposes in newborns. For neonatal testing, robust and minimally invasive specimens, such as DBSs, are required. Some data have applied MPS to DBSs to detect various diseases21 as well as WD.22 The approach of DNA-based screening of recurrent mutations followed by whole exon sequencing of ATP7B using MPS can therefore be used to screen high-risk or general populations. However, patients carrying mutations other than these recurrent mutations could be missed. Because of the rapid decline in the cost of sequencing in recent years, coding exon sequencing by MPS as a first tier screening test may be possible in the near future.
In summary, targeted screening of ATP7B in DBS samples revealed a higher mutant allele frequency than reported previously. Retrospective review of sequencing data of all ATP7B coding exons supported this finding. The discrepancy in disease prevalence between DNA-based estimation and clinical ascertainment suggests underdiagnosis of WD. Considering the limitations of diagnosing WD based on ceruloplasmin level, a tiered approach of screening of recurrent mutations followed by MPS could be a viable screening option for high-risk populations or for mass population screening.
References
Shah, A. B., Chernov, I., Zhang, H. T., Ross, B. M., Das, K., Lutsenko, S. et al. Identification and analysis of mutations in the Wilson disease gene (ATP7B): population frequencies, genotype-phenotype correlation, and functional analyses. Am. J. Hum. Genet. 61, 317–328 (1997).
Loudianos, G., Dessi, V., Lovicu, M., Angius, A., Nurchi, A., Sturniolo, G. C. et al. Further delineation of the molecular pathology of Wilson disease in the Mediterranean population. Hum. Mutat. 12, 89–94 (1998).
Kim, E. K., Yoo, O. J., Song, K. Y., Yoo, H. W., Choi, S. Y., Cho, S. W. et al. Identification of three novel mutations and a high frequency of the Arg778Leu mutation in Korean patients with Wilson disease. Hum. Mutat. 11, 275–278 (1998).
Yoo, H. W. Identification of novel mutations and the three most common mutations in the human ATP7B gene of Korean patients with Wilson disease. Genet. Med. 4, 43S–48S (2002).
Park, S., Park, J. Y., Kim, G. H., Choi, J. H., Kim, K. M., Kim, J. B. et al. Identification of novel ATP7B gene mutations and their functional roles in Korean patients with Wilson disease. Hum. Mutat. 28, 1108–1113 (2007).
Dong, Y., Ni, W., Chen, W. J., Wan, B., Zhao, G. X., Shi, Z. Q. et al. Spectrum and classification of ATP7B variants in a large cohort of Chinese patients with Wilson's disease guides genetic diagnosis. Theranostics 6, 638–649 (2016).
Shimizu, N., Kawase, C., Nakazono, H., Hemmi, H., Shimatake, H. & Aoki, T. A novel RNA splicing mutation in Japanese patients with Wilson disease. Biochem. Biophys. Res. Commun. 217, 16–20 (1995).
Okada, T., Shiono, Y., Hayashi, H., Satoh, H., Sawada, T., Suzuki, A. et al. Mutational analysis of ATP7B and genotype–phenotype correlation in Japanese with Wilson's disease. Hum. Mutat. 15, 454–462 (2000).
Zappu, A., Magli, O., Lepori, M. B., Dessi, V., Diana, S., Incollu, S. et al. High incidence and allelic homogeneity of Wilson disease in 2 isolated populations: a prerequisite for efficient disease prevention programs. J. Pediatr. Gastroenterol. Nutr. 47, 334–338 (2008).
Kim, G. H., Yang, J. Y., Park, J. Y., Lee, J. J., Kim, J. H. & Yoo, H. W. Estimation of Wilson's disease incidence and carrier frequency in the Korean population by screening ATP7B major mutations in newborn filter papers using the SYBR green intercalator method based on the amplification refractory mutation system. Genet. Testing 12, 395–399 (2008).
Park, H. D., Ki, C. S., Lee, S. Y. & Kim, J. W. Carrier frequency of the R778L, A874V, and N1270S mutations in the ATP7B gene in a Korean population. Clin. Genet. 75, 405–407 (2009).
Song, M. J., Lee, S. T., Lee, M. K., Ji, Y., Kim, J. W. & Ki, C. S. Estimation of carrier frequencies of six autosomal-recessive Mendelian disorders in the Korean population. J. Hum. Genet. 57, 139–144 (2012).
Dong, C., Wei, P., Jian, X., Gibbs, R., Boerwinkle, E., Wang, K. et al. Comparison and integration of deleteriousness prediction methods for nonsynonymous SNVs in whole exome sequencing studies. Hum. Mol. Genet. 24, 2125–2137 (2015).
Coffey, A. J., Durkie, M., Hague, S., McLay, K., Emmerson, J., Lo, C. et al. A genetic study of Wilson's disease in the United Kingdom. Brain 136, 1476–1487 (2013).
Ala, A., Walker, A. P., Ashkan, K., Dooley, J. S. & Schilsky, M. L. Wilson's disease. Lancet 369, 397–408 (2007).
Hahn, S. H., Lee, S. Y., Jang, Y. J., Kim, S. N., Shin, H. C., Park, S. Y. et al. Pilot study of mass screening for Wilson's disease in Korea. Mol. Genet. Metab. 76, 133–136 (2002).
Nakayama, K., Kubota, M., Katoh, Y., Sawada, Y., Saito, A., Nishimura, K. et al. Early and presymptomatic detection of Wilson's disease at the mandatory 3-year-old medical health care examination in Hokkaido Prefecture with the use of a novel automated urinary ceruloplasmin assay. Mol. Genet. Metab. 94, 363–367 (2008).
Kroll, C. A., Ferber, M. J., Dawson, B. D., Jacobson, R. M., Mensink, K. A., Lorey, F. et al. Retrospective determination of ceruloplasmin in newborn screening blood spots of patients with Wilson disease. Mol. Genet. Metab. 89, 134–138 (2006).
Owada, M., Suzuki, K., Fukushi, M., Yamauchi, K. & Kitagawa, T. Mass screening for Wilson's disease by measuring urinary holoceruloplasmin. J. Pediatr. 140, 614–616 (2002).
Hahn, S. H. Population screening for Wilson's disease. Ann. NY Acad. Sci. 1315, 64–69 (2014).
Lefterova, M. I., Shen, P., Odegaard, J. I., Fung, E., Chiang, T., Peng, G. et al. Next-generation molecular testing of newborn dried blood spots for cystic fibrosis. J. Mol. Diagn. 18, 267–282 (2016).
Nemeth, D., Arvai, K., Horvath, P., Kosa, J. P., Tobias, B., Balla, B. et al. Clinical use of next-generation sequencing in the diagnosis of Wilson's disease. Gastroenterol. Res. Pract. 2016, 4548039 (2016).
Acknowledgements
This study was supported by a grant from the Korean Ministry of Trade, Industry and Energy (10053626).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Jang, JH., Lee, T., Bang, S. et al. Carrier frequency of Wilson’s disease in the Korean population: a DNA-based approach. J Hum Genet 62, 815–818 (2017). https://doi.org/10.1038/jhg.2017.49
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2017.49
This article is cited by
-
Carrier frequency estimation of pathogenic variants of autosomal recessive and X-linked recessive mendelian disorders using exome sequencing data in 1,642 Thais
BMC Medical Genomics (2024)
-
Prevalence of common autosomal recessive mutation carriers in women in the Southern Vietnam following the application of expanded carrier screening
Scientific Reports (2024)
-
A Study of Dopaminergic Pathway in Neurologic Wilson Disease with Movement Disorder
Molecular Neurobiology (2023)
-
The global prevalence of Wilson disease from next-generation sequencing data
Genetics in Medicine (2019)
-
Wilson disease missense mutations in ATP7B affect metal-binding domain structural dynamics
BioMetals (2019)