Abstract
Intra-tumor heterogeneity is a big barrier to precision medicine. To explore the underlying clonal diversity in lung adenocarcinomas, we selected nine individuals with whole-genome sequencing data from primary and matched metastatic tumors as a cohort for study. Similar global pattern of arm-level copy number changes and large variations of somatic single-nucleotide variant between the primary and metastasis are observed in the majority of cases. Importantly, we found breakage-fusion-bridge (BFB) cycles acting as an important mechanism for underlying cancer gene amplification, such as amplification of CDK4, CDKN3 and FGFR1 in early stage. We also identified recurrent focal amplification of gene CCNY derived from BFB in two metastatic tumors, but not in primary tumor. Clonal analysis of case 236T demonstrated that mutational processes are varying with tumor progression. Collectively, our data provide new insights into genetic diversity and potential therapeutic target in lung adenocarcinoma.
Similar content being viewed by others
Introduction
Lung cancer is the most lethal cancer in China and all over the world. Nearly 40% of all lung cancers are adenocarcinoma. Around 50% lung cancers are diagnosed with lymph node invasion and distant organ metastasis. Target therapy regarding to druggable alterations such as ALK-EML4 fusion, EGFR and KRAS mutations are widely used for individuals with lung adenocarcinoma.1, 2, 3 However, intra-tumor heterogeneity has posted a great challenge to therapy as many tumors begin to relapse and are resistant to therapeutic drugs after remission for several months.4 Clarifying the evolutionary processes operative in oncogenic path that lead to clonal diversity, especially for metastatic clone, is essential for targeted therapy in lung adenocarcinoma.
Previously, genomic studies of lung adenocarcinoma focused on driver genes identification and clonal evolution based on exome-sequencing and single-nucleotide polymorphism array.5, 6 However, studies on genomic heterogeneity based on whole-genome sequencing that enable us to detect somatic structural variations are relatively limited. Hence, we used the whole genomic sequencing data of primary and matched metastatic lung adenocarcinoma from published data as a cohort for study. Recurrent focal amplification of CCNY derived by breakage-fusion-bridge (BFB) cycle is found in two metastatic tumors. We also elucidate the high concordance of copy number profiling in eight cases, meanwhile the remaining one case that exhibited significant differences of genomic alterations might be due to genome doubling.
Materials and Methods
Sample introduction
All 24 cases with whole-genome sequencing data of primary and matched metastatic lung adenocarcinoma were from published data,7 which have been deposited to the European Genome-phenome Archive under accession number EGAS00001000982. Of these, 15 cases having low tumor content in either primary or metastatic tumors were removed.
Somatic mutations calling and annotation
We used BWA (v0.5.9, http://bio-bwa.sourceforge.net/) with default parameters to align sequencing reads to the NCBI human reference genome (hg19). GATK are used to realign the mapped reads. The somatic single-nucleotide variants were subsequently called by MuTect8 using paired alignment files of tumor and normal genomes as input. Somatic mutations having at least four supporting variant reads are retained and further annotated by Oncotator (v1.8.0.0).9
Somatic structural variations
We used Meerkat10 to predict somatic structural variants and their breakpoints with suggested parameters. Candidate breakpoints were first found based on soft-clipped and split reads, and then confirmed to be precise breakpoints by local alignments. Mutational mechanisms were predicted based on homology and sequencing features. After filtering out germline events and other artifacts, we got the final somatic structural variants and further generated fusion genes.
Copy number change
We used Patchwork11 to identify copy number alterations from nine primary and metastatic tumors. Segmentation results were then used for the subsequent analysis.
Break-fusion-bridge detection
We inferred BFB events from the Meerkat results based on three criteria proposed by Peter Campbell et al:12 (1) Inversion is single inverted that is read as invers_f or invers_r by Meerkat. (2) Inversion segments have copy number change versus the adjacent position. (3) The two ends of the inversions must be separated by <30 kb.
Cancer cell fraction estimation and genome doubling prediction
We applied ABSOLUTE13 to transform relative copy number segments into absolute copy number for primary and matched metastatic tumors. Cancer cell fraction is estimated based on local copy number and variant allele frequency of each somatic single-nucleotide variant (SNV). Somatic SNVs are defined as sub-clonal mutations if they have >=50% probability of being sub-clonal mutations.
Results
Genomic diversity within intra-individual genomes
We applied ABSOLUTE13 to quantify copy number change profiled by Patchwork from 24 primary and metastatic tumors, of which nine cases have comparable tumor contents between primary and metastatic tumors (Supplementary Table 1). The remaining 15 cases exhibit nearly no copy number changes in either primary or metastatic tumors, suggesting low proportion of cancer cells in these samples. Therefore, we focused on nine cases with high neoplastic purities that are feasible to characterize intra-tumor heterogeneity in lung adenocarcinoma.
Based on copy number profiling, we found the genomes of 8 cases are nearly diploid except case 236T. All cases shared similar global pattern of arm-level change between primary tumor and metastases (Supplementary Figure 1). Of these eight cases, gene-level copy number correlation ranges from 0.54 to 0.89. However, we still found sporadic distinct arm-level change: gain of chr2 is specific to primary tumor in case 206T; gain of chr4 occurred specifically in metastasis 155T (Supplementary Figure 1). It is worth noting that 4q gain is a metastatic signature in lung cancer.14 This data suggests limited copy number change of intra-individual genomes in lung adenocarcinoma.
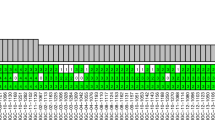
In contrast, the number of common somatic SNVs varied between primary tumors and metastases and nearly half of mutations are specific to primary genome or metastases (Figure 1a). The proportion of sub-clonal mutation varied across nine cases, of which three case including 236T, 155T and 206T have more sub-clonal mutations in metastasis compared to primary tumor (Figure 1b). Interestingly, TP53 and PIK3CA mutations aresepeartely found in primary tumors 23T and 236T, while MET has two missense mutations in metastatic cases 075T and 155T (Figure 1c).
The commonality and difference in nine pairs of matched primary and metastasis lung cancer samples. (a) The common and unique mutation in coding area between primary and metastasis samples. (b) Subclone mutation rate in primary and metastasis. Each square represents a sample, x and y scale represent the mutation rate in primary and metastasis, black number marked represent sample name. (c) Number of coding mutations in primary and metastasis. M indicated metastasis and P indicated primary. (d) Driver mutation events in nine matched primary and metastasis samples including somatic mutation, focal amplification or deletion, fusion and BFB. Each column represents a primary (LB*) or metastasis (LC*) sample. A full color version of this figure is available at the Journal of Human Genetics journal online.
Breakage-fusion-bridge in primary and metastatic genomes
We analyzed somatic structural variant between primary and metastasis. Great diversity of structural variations was observed across nine cases (Supplementary Figure 2 and Supplementary Table 2), albeit they shared similar mutational mechanisms between primary tumor and metastases. Of note, case 075T has large number of structural variations (Supplementary Figure 2) indicating high genomic instability. In addition, EGFR is amplified in 67% samples and ALK-EML4 fusion was found in sample 156 (Figure 1d).
Some of the somatic structural variants were recognized as a single inverted rearrangement exhibiting layers of copy number amplification. These rearrangements may be termed ‘fold-back inversions’ as previous studies supported,12 which is most likely to be generated by BFB cycle. In total, we identified 154 fold-back inversions in primary tumors, as well as 83 in metastases. We observed 72% of fold-back inversions in primary tumors and 75% in metastases are mediated by micro-homology (Supplementary Figure 3).
Notably, we observed multiple cancer genes involved in BFB cycle. High-level of CDK4, CDKN3 and FGFR1 amplification derived from BFB cycle could be observed in both primary and metastatic tumors (Figures 2a–c), implying these BFB events occurred early in tumor development before tumor-cell dissemination. CDK4 encodes a Ser/Thr protein that is important for cell cycle G1 phase, amplification of CDK4 gene has been found in various human tumors including lung tumors, which can lead to tumorigenesis.15 CDKN3 is a cyclin-dependent kinase inhibitor, and this gene was reported to be overexpressed in most human cancer tissues and may have association with poor survival in lung cancer.16 FGFR1 amplification is common in lung and other human cancer and it was predicted as a predictive prognostic maker.17 Interestingly, we found recurrent CCNY BFB-derived amplification specifically in metastatic lung cancer (Figure 2d). CCNY is a member of cyclins controlled cell division cycles and regulated cyclin-dependent kinases. Overexpression of CCNY can result in tumor proliferation, and has been reported to be a therapeutic target in non-small-cell lung cancer.18 However, the underlying mechanism of CCNY amplification in metastatic lung cancer has not been elucidated before. Further functional study is needed to recapitulate its role in metastases. Collectively, our results confirmed an important role of BFB cycles derived gene amplifications in lung cancer, and targeted therapy may benefit lung cancer patients with these amplified oncogenes.
BFB events across nine LC paired primary and metastases. The upper panel shows the intra- rearrangements in each chromosome. The bottom panel shows the normalized coverage. (a) CDK4 amplification. (b) CDKN3 amplification. (c) FGFR1 amplification. (d) CCNY amplification. A full color version of this figure is available at the Journal of Human Genetics journal online.
Branched evolution in case 236T
Even though high concordant genomic alterations were found in most primary and metastatic genomes, an exception was found in patient 236, wherein a large difference of arm-level change was observed with 15% common SNVs among them. Clustering with arm level copy number variant change showed that primary sample 236T and metastasis sample 236C had a much greater genetic evolution distance which confirmed the tremendous genomic differences between them (Supplementary Figure 4). Specifically, metastatic genome was predicted to undergo genome doubling using ABSOLUTE that provides reasonable explanation for large genomic changes from primary tumor. To further explore the characteristics of mutational processes, we further inspected its genomic evolution in detail.
During the study, ABSOLUTE13 was employed to estimate the cancer cell fraction in both somatic copy number aberration and somatic single nucleotide variant (Figure 3c). On the basis of the cancer cell fraction of each mutation, the mutations were classified into three categories: early, middle and late (Figure 3b). It was presumed that the clonal mutation belonged to the early events and sub-clonal mutation belonged to the late events. Mutations in the clonal group were further classified into mutations occurring before metastasis (mutations appear in both primary and metastasis) and mutations that occur after metastasis (mutations appear only in primary or metastasis). Those mutations that happen after metastasis were defined as middle events and the mutations that occurred before metastasis were considered to be early events. Hence in our data, it was observed that primary tumor and metastases of the case 236 and other eight cases were all derived from a common ancestor clone, and then each slowly evolves into two independent clones.
The evolutionary processes in sample 236T. (a) Absolute copy number profiling of sample 236 indicating genome-doubling event in metastatic tumor. Blue, purple and red represent deletion, dipoid and amplification, respectively. (b) Inferred phylogenetic tree. (c) Distribution of cancer cell fraction. Red, blue and purple represent different stages of evolution mentioned in the article. The pie shows different mutation types. A full color version of this figure is available at the Journal of Human Genetics journal online.
Also, we compared the copy number changes in case 236 (Figure 3a). The 3p deletion was present in both primary and metastasis, indicating it belonged to an early event. Further, the 3p deletion that frequently occurred in lung cancer has been reported to influence the telomerase activity and accelerate the tumor evolution.19 Similar partial deletions in 5q and 6q were present in primary and metastasis. Amplification of chr17q and partial chr19 was observed whereas the whole genome-doubling event occurred only during metastasis. The relative late genome-doubling event in metastasis, which caused many copy number loss and gain has drastically altered the metastatic genomic structure leading to an immense genomic divergence with primary cancer.
We aslo analyzed the non-synonymous mutations in this sample, and found that LRP1B (p.K2623N) and KMT2C (p.R380L, p.C391*) shared missense mutations in both primary and metastatic tumors (Figure 3b). LRP1B frequently mutated in lung adenocarcinoma and it could be associated with lung carcinogenesis.7 Besides, the PIK3CA H1047R mutation which has been reported in COSMIC (v62) was also observed in LB236. PIK3CA is a well-established oncogene in lung adenocarcinoma, and this mutation can increase the downstream kinase activity, decrease cell apoptosis and promote tumor-cell invasion. Further, recent studies showed that PIK3CA mutation tends to emerge in sub-clones associated with the APOBEC signature environment.20 In consistent, H1047R mutation was a later event specific to primary tumor in our data. Moreover, it was observed that mutational signatures evolved along with tumor progression in primary tumor. The percentage of C>A mutation decreased in primary tumor, contrast to the increased percentage of C>T and C>G mutation, especially for C>T and C>G mutations at TpC di-nucleotides (Supplementary Figure 5). Previous reports showed that smoking can increase the percentage of C>A mutation, and the evolution of mutation spectrum in our data indicate that the mutation burden caused by smoking might decrease, whereas mutation burden induced by APOBEC increase21 in primary tumor. Similar change of mutation spectrum was observed in metastasis, suggesting that similar mutational mechanisms in later progression of primary tumor and metastasis.
Discussion
In summary, in the process of this study we have demonstrated the genomic diversity between primary and metastatic tumors. As arm-level change is relatively stable with few copy number changes during genome evolution, somatic SNV are largely differentiated in the majority of cases. These findings suggest differential evolutionary rate in different time points for copy number change and somatic mutations.
Chromosomal instability is defined as continual loss or gain of whole chromosome or sub-chromosome and numerical chromosome instability and structural instability.22 Mostly, CIN is associated with multi-drug responses and poor prognosis.23, 24 Genome doubling have the ability to increase genome instability in a cancer genome.25 Based on our data, we found genome doubling occurred specifically in the metastasis of case 023, and also uncovered recurrent BFB-derived amplification of gene CCNY specific to metastases. The biological role of genome doubling and BFB events in metastasis needs further investigation.
Finally, we demonstrated the branched evolutionary process through representative case 023T. The data showed most of mutations occurred in middle or later stage with PIK3CA hotspot mutation in middle stage. These results are consistent with intra-tumor heterogeneity report in kidney cancer that most of the driver or passenger mutations occurred in branched or private stage, and not in the founding clone. Besides, a change of mutational mechanisms was observed with tumor progression. These results indicated that distinct functions of driver mutations were involved in cancer initiation and progression stage.
References
Kwak, E. & Bang, Y. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N. Engl. J. Med. 363, 1693–1703 (2010).
Linardou, H., Dahabreh, I. J., Kanaloupiti, D., Siannis, F., Bafaloukos, D., Kosmidis, P. et al. Assessment of somatic k-RAS mutations as a mechanism associated with resistance to EGFR-targeted agents: a systematic review and meta-analysis of studies in advanced non-small-cell lung cancer and metastatic colorectal cancer. Lancet Oncol. 9, 962–972 (2008).
Shaw, A. T., Yeap, B. Y., Mino-Kenudson, M., Digumarthy, S. R., Costa, D. B., Heist, R. S. et al. Clinical features and outcome of patients with non-small-cell lung cancer who harbor EML4-ALK. J. Clin. Oncol. 27, 4247–4253 (2009).
Sequist, L. V., Waltman, B. a., Dias-Santagata, D., Digumarthy, S., Turke, A. B., Fidias, P. et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci. Transl. Med. 3, 75ra26 (2011).
Collisson, E. A., Campbell, J. D., Brooks, A. N., Berger, A. H., Lee, W., Chmielecki, J. et al. Comprehensive molecular profiling of lung adenocarcinoma. Nature 511, 543–550 (2014).
Pelosi, G., Pellegrinelli, A., Fabbri, A., Tamborini, E., Perrone, F., Settanni, G. et al. Deciphering intra-tumor heterogeneity of lung adenocarcinoma confirms that dominant, branching, and private gene mutations occur within individual tumor nodules. Virchows Arch. 486, 651–662 (2016).
Wu, K., Zhang, X., Li, F., Xiao, D., Hou, Y., Zhu, S. et al. Frequent alterations in cytoskeleton remodelling genes in primary and metastatic lung adenocarcinomas. Nat. Commun. 6, 10131 (2015).
Cibulskis, K., Lawrence, M. S., Carter, S. L., Sivachenko, A., Jaffe, D., Sougnez, C. et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 31, 213–219 (2013).
Ramos, A. H., Lichtenstein, L., Gupta, M., Lawrence, M. S., Pugh, T. J., Saksena, G. et al. Oncotator: cancer variant annotation tool. Hum. Mutat. 36, E2423–E2429 (2015).
Yang, L., Luquette, L. J., Gehlenborg, N., Xi, R., Haseley, P. S., Hsieh, C. H. et al. Diverse mechanisms of somatic structural variations in human cancer genomes. Cell 153, 919–929 (2013).
Mayrhofer, M., DiLorenzo, S. & Isaksson, A. Patchwork: allele-specific copy number analysis of whole-genome sequenced tumor tissue. Genome Biol. 14, R24 (2013).
Campbell, P. J., Yachida, S., Mudie, L. J., Stephens, P. J., Pleasance, E. D., Stebbings, L. A. et al. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature 467, 1109–1113 (2010).
Carter, S. L., Cibulskis, K., Helman, E., McKenna, A., Shen, H., Zack, T. et al. Absolute quantification of somatic DNA alterations in human cancer. Nat. Biotechnol. 30, 413–421 (2012).
Liu, H., Park, J., Manning, C., Goehlmann, H. W. H. & Marshall, D. J. Metastatic signature in lung cancer is associated with sensitivity to anti-integrin αV monoclonal antibody intetumumab. Genes Chromosom. Cancer 53, 349–357 (2014).
Wikman, H., Nymark, P., Väyrynen, A., Jarmalaite, S., Kallioniemi, A., Salmenkivi, K. et al. CDK4 is a probable target gene in a novel amplicon at 12q13.3–q14.1 in lung cancer. Genes Chromosom. Cancer 42, 193–199 (2005).
Fan, C., Chen, L., Huang, Q., Shen, T., Welsh, E. A., Teer, J. K. et al. Overexpression of major CDKN3 transcripts is associated with poor survival in lung adenocarcinoma. Br. J. Cancer 113, 1735–1743 (2015).
Zhao, H., Ning, L. L., Wang, Z. Y., Li, H. T., Qiao, D., Yao, Y. et al. Calcitonin gene-related peptide inhibits osteolytic factors induced by osteoblast in co-culture system with breast cancer. Cell Biochem. Biophys. 70, 1097–1104 (2014).
Yue, W., Zhao, X., Zhang, L., Xu, S., Liu, Z., Ma, L. et al. Cell cycle protein cyclin Y is associated with human non-small-cell lung cancer proliferation and tumorigenesis. Clin. Lung Cancer 12, 43–50 (2011).
Iniesta, P., Gonzalez-Quevedo, R., Moran, A., Garcia-Aranda, C., de Juan, C., Sanchez-Pernaute, A. et al. Relationship between 3p deletions and telomerase activity in non-small-cell lung cancer: prognostic implications. Br. J. Cancer 90, 1983–1988 (2004).
Mcgranahan, N., Favero, F., De Bruin, E. C., Birkbak, N. J., Szallasi, Z. & Swanton, C. Clonal status of actionable driver events and the timing of mutational processes in cancer evolution. Sci. Transl. Med. 7, 283ra54–283ra54 (2015).
de Bruin, E. C., McGranahan, N., Mitter, R., Salm, M., Wedge, D. C., Yates, L. et al. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science 346, 251–256 (2014).
Janssen, A. & Medema, R. H. Genetic instability: tipping the balance. Oncogene 32, 4459–4470 (2013).
McGranahan, N., Burrell, R. A., Endesfelder, D., Novelli, M. R. & Swanton, C. Cancer chromosomal instability: therapeutic and diagnostic challenges. EMBO Rep. 13, 528–538 (2012).
Lee, A. J. X., Endesfelder, D., Rowan, A. J., Walther, A., Birkbak, N. J., Futreal, P. A. et al. Chromosomal instability confers intrinsic multidrug resistance. Cancer Res. 71, 1858–1870 (2011).
Dewhurst, S. M., McGranahan, N., Burrell, R. A., Rowan, A. J., Grönroos, E., Endesfelder, D. et al. Tolerance of whole-genome doubling propagates chromosomal instability and accelerates cancer genome evolution. Cancer Discov. 4, 175–185 (2014).
Acknowledgements
We thank all of the patients and families that contributed to this study. This work is supported by the funding from the Jilin Department of Health (3D514CQ13430). This work is supported by grants of Guangzhou Health-Medical Collaborative Innovation Project (NO. 201508020247). We also thank The First Affiliated Hospital of Guangzhou Medical University for sharing the whole-genome data.
Author contributions
BY and YZ designed and managed the research. WL and CY performed the bioinformatics analysis. YZ, WL, CY, TX and XM wrote the manuscript. XL, XZ and WZ revised the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Yang, B., Luo, L., Luo, W. et al. The genomic dynamics during progression of lung adenocarcinomas. J Hum Genet 62, 783–788 (2017). https://doi.org/10.1038/jhg.2017.40
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2017.40