Abstract
Cerebellar atrophy is recognized in various types of childhood neurological disorders with clinical and genetic heterogeneity. Genetic analyses such as whole exome sequencing are useful for elucidating the genetic basis of these conditions. Pathological recessive mutations in Sep (O-phosphoserine) tRNA:Sec (selenocysteine) tRNA synthase (SEPSECS) have been reported in a total of 11 patients with pontocerebellar hypoplasia type 2, progressive cerebellocerebral atrophy or progressive encephalopathy, yet detailed clinical features are limited to only four patients. We identified two new families with progressive cerebellar atrophy, and by whole exome sequencing detected biallelic SEPSECS mutations: c.356A>G (p.Asn119Ser) and c.77delG (p.Arg26Profs*42) in family 1, and c.356A>G (p.Asn119Ser) and c.467G>A (p.Arg156Gln) in family 2. Their development was slightly delayed regardless of normal brain magnetic resonance imaging (MRI) in infancy. The progression of clinical symptoms in these families is evidently slower than in previously reported cases, and the cerebellar atrophy milder by brain MRI, indicating that SEPSECS mutations are also involved in milder late-onset cerebellar atrophy.
Similar content being viewed by others
Introduction
Cerebellar atrophy or hypoplasia is recognized in various types of childhood neurological disorders with clinical and genetic heterogeneity. Atrophy and hypoplasia are both strictly characterized: cerebellar atrophy describes a loss of cerebellar tissue with an initially normal structure, whereas cerebellar hypoplasia is defined by a compact cerebellum of reduced volume that does not fill the posterior fossa.1 However, hypoplasia is often included in atrophy because atrophy is often difficult to be distinguished from hypoplasia.
Cerebellar atrophy is classified into congenital and postnatally acquired cerebellar atrophies. Congenital cerebellar atrophies include several genetic disorders such as pontocerebellar hypoplasia (PCH), spinocerebellar ataxia and inherited metabolic diseases.1 Pontocerebellar hypoplasia is a clinically and genetically heterogeneous group of inherited neurodevelopmental disorders predominantly characterized by prenatal onset of stunted growth and decay of cerebral structures. The initial classification is based on two subtypes, PCH1 and PCH2, and is determined from neuropathological findings, specifically the presence or absence of anterior horn cell degeneration.2 Postnatally acquired cerebellar atrophies are caused by cerebral palsy with severe perinatal asphyxia, congenital infection and exposure to teratogens.
Sep (O-phosphoserine) tRNA:Sec (selenocysteine) tRNA synthase (SEPSECS) encodes the SepSecS protein that is important for synthesis of selenocysteine, a selenoprotein component that is essential for mammalian brain development. Pathological recessive SEPSECS mutations have been reported in a total of 11 patients with PCH type 2, progressive cerebellocerebral atrophy or progressive encephalopathy, although detailed clinical features are limited to only four patients. Most patients had exhibited a severe neurodevelopment disorder since their infantile periods.3, 4
In this study, we describe two families with cerebellar atrophy that harbor biallelic SEPSECS mutations. Detailed clinical information is described and compared with previously reported cases.
Materials and methods
Patients
A total of 96 families with cerebellar atrophy (including 85 previously described families) were analyzed.5 Both static and progressive cerebellar atrophy were included. Detailed clinical information was obtained from the clinicians who examined the patients. The institutional review board of Yokohama City University of Medicine approved the experimental protocols. Informed consent was obtained for all patients, in agreement with Japanese regulation requirements.
Whole exome sequencing
Genomic DNA was isolated from peripheral blood leukocytes using QuickGene 610L (Wako, Osaka, Japan), then captured using the SureSelect Human All Exon v4 or v5 Kit (51 Mb; Agilent Technologies, Santa Clara, CA, USA), and sequenced on an Illumina HiSeq2000 or Hiseq2500 (Illumina, San Diego, CA, USA) with 101-bp paired-end reads. Exome data processing, variant calling and variant annotation were performed as previously described.6 Common single-nucleotide polymorphisms (SNPs) with minor allele frequencies ⩾1% in dbSNP 135 and variants observed in >5 of our in-house 575 control exomes database were filtered out. Among the remaining rare variants, we focused on amino acid altering or splicing-affecting variants. Particular attention was given to mutations in previously reported causative genes associated with cerebellar atrophy. SEPSECS mutations were validated by Sanger sequencing using genomic DNA from patients and their parents as a template.
Results
Genetic analysis
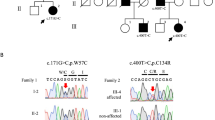
Using whole exome sequencing, we identified potential biallelic mutations in SEPSECS (NM_016955.3) in two individuals from two families: c.356A>G (p.Asn119Ser) and c.77delG (p.Arg26Profs*42) in individual 1, and c.356A>G (p.Asn119Ser) and c.467G>A (p.Arg156Gln) in individual 2 (Figure 1a). These compound heterozygous mutations were verified by Sanger sequencing using patient and parental samples. The amino acids affected by two missense mutations were conserved in eukaryotic SepSecS proteins (Figure 1b) and located on the SepSecS domain (Figure 1c). These mutations were absent in dbSNP 138, our in-house database and the Exome Aggregation Consortium (ExAC) database, and are therefore likely to be extremely rare. Web-prediction tools (specifically, SIFT (http://sift.jcvi.org/), Pholyphen-2 (http://genetics.bwh.harvard.edu/pph2/) and MutationTaster (http://www.mutationtaster.org/)) indicated that both mutations are pathogenic (Supplementary Table 1).
(a) Familial pedigrees and SEPSECS mutations. (b) Electropherograms of mutations and evolutionary conservation of amino acids derived from the missense mutations in Caenorhabditis elegans to humans. (c) Schematic of the SEPSECS gene. Locations of mutations are depicted (upper, novel mutations identified here; lower: previously described mutations). SEPSECS, Sep (O-phosphoserine) tRNA:Sec (selenocysteine) tRNA synthase.
Clinical presentation
Clinical features of individuals with SEPSECS mutations are presented in Table 1. Individuals 1 and 2 showed similar clinical courses. During infancy, their development was slightly delayed and initial brain magnetic resonance imaging (MRI) showed normal findings (individual 1 at 6 months of age, and individual 2 at 5 years of age). Ataxia and motor disability slowly progressed. Cerebellar atrophy was first recognized by MRI at 9 years and 18 years of age, respectively.
Individual 1 was a 10-year-old girl, and the first child born to healthy non-consanguineous parents after an uncomplicated 41-week pregnancy. She had no dysmorphic features at birth, and acquired eye pursuits at 2 months of age. She was referred to our hospital at 3 months because of downward nystagmus. Neurological examination revealed that she was floppy but her reflexes were normal. Although her head circumference was within normal range at birth, her head growth was slow. Head circumference at 4 years was 47.0 cm (−2.1 s.d.). Brain MRI at 6 months of age was normal (Figure 2a). She was able to sit alone at 8 months, and her nystagmus disappeared by 9 months. She moved by bottom shuffling before walking. She walked with support and alone at 1 year and 3 months, and 3 years and 6 months, respectively. She was able to speak several meaningful words, but could not speak in sentences at 3 years and 6 months. Around the same time, rotating and horizontal nystagmus appeared and she had intention tremors and dysmetria.
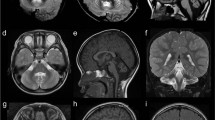
Brain magnetic resonance imaging (MRI) of individual 1 at 6 months (a) and 9 years (b–c) of age, and individual 2 at 18 years (d–f) of age. Axial (a, b) and sagittal (c) T2-weighted images show mild atrophy of the frontoparietal lobes and mild cerebellar atrophy at 9 years (b) of age, but not at 6 months (a) of age. Axial (d) and coronal (e) T2-weighted images show mild cerebral atrophy and cerebellar lobe atrophy, respectively. White matter around posterior horn of lateral ventricle showed slightly hyperintensity. Sagittal T1-weighted image (f) shows cerebellocerebral and vermis atrophy.
At 9 years of age, she was admitted to hospital because of respiratory infection. Brain MRI revealed progressive atrophy of the cerebellum and frontoparietal cerebral lobe (compared with 6 months) (Figures 2b and c). Electroencephalography, peripheral conduction study, cerebrospinal fluid examination and somatosensory evoked potentials were normal. Neurological examination revealed progressive spasticity and ataxia with an unstable walk, even using a walking aid.
Individual 2, a 21-year-old girl, was the first child born to healthy non-consanguineous parents. There was no familial history. Although she showed congenital microcephaly at birth, her initial development was normal: she obtained head control at 3 months of age. Thereafter, her motor development was slightly delayed but she could sit and pull to stand at 10 months, and 1 year and 6 months, respectively, but could not walk until 2 years.
She was able to speak meaningful words and sentences at 2 and 5 years of age, respectively. After she graduated from special support school (instead of regular senior high school), she attended our medical institution because she had difficulty in walking without support. Neurological examination revealed that she had ataxia, coordination disturbance, dysmetria, hypotonia and deep tendon hyperreflexia. Brain MRI revealed cerebellar and vermis atrophy and slight cerebral atrophy (Figures 2d–f).
Discussion
Previous studies have reported clinical features of patients with SEPSECS defects. Common symptoms among most patients, including the two described here, include developmental delay, hypotonia and cerebellar ataxia. Brain MRI frequently shows atrophy of the cerebellum, vermis and cerebrum. Nystagmus, microcephaly, seizure, spasticity and encephalopathy are also observed in some patients. Brain atrophy is recognized at various developmental stages, but mostly at birth to childhood. In our patients, cerebellar atrophy in individuals 1 and 2 was recognized at the age of 9 and 18 years, respectively. This is later than in previous cases, and the degree of atrophy is also milder. This indicates that the phenotype of SEPSECS defects includes milder cases than previously reported. In addition, the fact that our milder cases presented with microcephaly and developmental delay implied that SEPSECS plays important roles in brain development.
SEPSECS consists of 12 exons and encodes the SepSecS protein of 501 amino acids that catalyzes the last step in the conversion of Sep-tRNA to Sec-tRNA.7 This reaction is the sole route to selenocysteine biosynthesis in humans.8 Selenocysteine is a component of selenoproteins, and human selenoproteins include 25 members with biological functions implicated in diverse human diseases ranging from cardiovascular to immunoreactive disorders.9 Conditional Trsp knockout mice, in which neuronal selenoproteins are deficient, show cerebellar hypoplasia and Purkinje cell loss,10 suggesting that selenoproteins are essential for mammalian brain development. In addition, selenoproteins are suggested to play an important role in antioxidant defense.9 Decreasing selenoprotein synthesis may damage organs with high mitochondrial activity because mitochondria are one of the main sources of cellular reactive oxygen species.4 In a previous study, SEPSECS mutations caused clinical features similar to those of mitochondrial disease such as lactacidemia.4 Although blood and/or cerebrospinal fluid lactate levels were almost within the normal range in our two patients, possibly consistent with the milder phenotype, elevated lactates might be a key clinical feature in patients with SEPSECS mutations.
To date, five missense, one nonsense, one splice site change and a gross deletion have been reported in SEPSECS (Figure 1c and Table 2). Four patients (in three families) with compound heterozygous mutations (c.974C>G (p.Thr325Ser) and c.1287C>A (p.Tyr429*)) showed progressive encephalopathy with microcephaly and infantile epileptic seizures.4 Four patients with biallelic missense mutations (c.1001A>G (p.Tyr334Cys) and c.715G>A (p.Ala239Thr)) were diagnosed with progressive cerebellocerebral atrophy. In one of them, cerebral atrophy was recognized at the age of 18 months.3 A patient with a missense mutation (first methionine) and a splice site change (c.1A>G and c388+3A>G) showed PCH type 2D.11 In this study, individual 1 with missense and frameshift mutations showed late-onset progressive cerebellocerebral atrophy, whereas individual 2 had biallelic missense mutations and showed markedly late-onset progressive cerebellocerebral atrophy. This suggests that the genotypes do not clearly indicate phenotypic difference and severity. Considering the common mutation (c.356A>G (p.Asn119Ser)) in our two individuals, it is possible that c.77delG (p.Arg26Profs*42) (individual 1) has a more deleterious effect on SEPSECS function compared with c.467G>A (p.Arg156Gln) (individual 2), supporting the fact that individual 2 shows a much milder phenotype than individual 1.
In conclusion, we have identified two families with cerebellar atrophy arising from biallelic novel SEPSECS mutations. Late-onset and milder phenotypes were recognized, indicating that SEPSECS mutant phenotypes show a wide range of clinical phenotypes.
References
Poretti, A., Wolf, N. I. & Boltshauser, E. Differential diagnosis of cerebellar atrophy in childhood. Eur. J. Paediatr. Neurol. 12, 155–167 (2008).
Rudnik-Schoneborn, S., Barth, P. G. & Zerres, K. Pontocerebellar hypoplasia. Am. J. Med. Genet. C Semin. Med. Genet. 166C, 173–183 (2014).
Agamy, O., Ben Zeev, B., Lev, D., Marcus, B., Fine, D., Su, D. et al. Mutations disrupting selenocysteine formation cause progressive cerebello-cerebral atrophy. Am. J. Hum. Genet. 87, 538–544 (2010).
Anttonen, A. K., Hilander, T., Linnankivi, T., Isohanni, P., French, R. L., Liu, Y. et al. Selenoprotein biosynthesis defect causes progressive encephalopathy with elevated lactate. Neurology 85, 306–315 (2015).
Ohba, C., Haginoya, K., Osaka, H., Kubota, K., Ishiyama, A., Hiraide, T. et al. De novo KIF1A mutations cause intellectual deficit, cerebellar atrophy, lower limb spasticity and visual disturbance. J. Hum. Genet. 60, 739–742 (2015).
Saitsu, H., Nishimura, T., Muramatsu, K., Kodera, H., Kumada, S., Sugai, K. et al. De novo mutations in the autophagy gene WDR45 cause static encephalopathy of childhood with neurodegeneration in adulthood. Nat. Genet. 45, 445–449 449e441 (2013).
Palioura, S., Sherrer, R. L., Steitz, T. A., Soll, D. & Simonovic, M. The human SepSecS-tRNASec complex reveals the mechanism of selenocysteine formation. Science 325, 321–325 (2009).
Yuan, J., Palioura, S., Salazar, J. C., Su, D., O’Donoghue, P., Hohn, M. J. et al. RNA-dependent conversion of phosphoserine forms selenocysteine in eukaryotes and archaea. Proc. Natl Acad. Sci. USA 103, 18923–18927 (2006).
Bellinger, F. P., Raman, A. V., Reeves, M. A. & Berry, M. J. Regulation and function of selenoproteins in human disease. Biochem. J. 422, 11–22 (2009).
Wirth, E. K., Bharathi, B. S., Hatfield, D., Conrad, M., Brielmeier, M. & Schweizer, U. Cerebellar hypoplasia in mice lacking selenoprotein biosynthesis in neurons. Biol. Trace Elem. Res. 158, 203–210 (2014).
Zhu, X., Petrovski, S., Xie, P., Ruzzo, E. K., Lu, Y. F., McSweeney, K. M. et al. Whole-exome sequencing in undiagnosed genetic diseases: interpreting 119 trios. Genet. Med. 17, 774–781 (2015).
Makrythanasis, P., Nelis, M., Santoni, F. A., Guipponi, M., Vannier, A., Bena, F. et al. Diagnostic exome sequencing to elucidate the genetic basis of likely recessive disorders in consanguineous families. Hum. Mutat. 35, 1203–1210 (2014).
Alazami, A. M., Patel, N., Shamseldin, H. E., Anazi, S., Al-Dosari, M. S., Alzahrani, F. et al. Accelerating novel candidate gene discovery in neurogenetic disorders via whole-exome sequencing of prescreened multiplex consanguineous families. Cell Rep. 10, 148–161 (2015).
Acknowledgements
We thank the individuals and their families for their participation in this study. We also thank Nobuko Watanabe and Mai Sato for their excellent technical assistance. This work is supported in part by a grant for Research on Measures for Intractable Diseases (14525125); a grant for Comprehensive Research on Disability Health and Welfare (13802019); the Strategic Research Program for Brain Science (SRPBS) (11105137) and Practical Research Project for Rare/Intractable Diseases (27280301) from Japan Agency for Medical Research and Development; a Grant-in-Aid for Scientific Research on Innovative Areas (Transcription Cycle) (24118007) from the Ministry of Education, Culture, Sports, Science and Technology of Japan; Grants-in-Aid for Scientific Research (B) (25293085, 25293235) and (A) (13313587), challenging Exploratory Research (26670505) and Young Scientists (B) (26860816) from the Japan Society for the Promotion of Science; the fund for Creation of Innovation Centers for Advanced Interdisciplinary Research Areas Program in the Project for Developing Innovation Systems (11105305) from the Japan Science and Technology Agency; and the Takeda Science Foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Iwama, K., Sasaki, M., Hirabayashi, S. et al. Milder progressive cerebellar atrophy caused by biallelic SEPSECS mutations. J Hum Genet 61, 527–531 (2016). https://doi.org/10.1038/jhg.2016.9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2016.9
This article is cited by
-
Broadening the phenotype and genotype spectrum of novel mutations in pontocerebellar hypoplasia with a comprehensive molecular literature review
BMC Medical Genomics (2024)
-
Analysis of the Clinical Features and Imaging Findings of Pontocerebellar Hypoplasia Type 2D Caused by Mutations in SEPSECS Gene
The Cerebellum (2022)
-
Entire FGF12 duplication by complex chromosomal rearrangements associated with West syndrome
Journal of Human Genetics (2019)
-
What’s new in pontocerebellar hypoplasia? An update on genes and subtypes
Orphanet Journal of Rare Diseases (2018)
-
A novel PGAP3 mutation in a Croatian boy with brachytelephalangy and a thin corpus callosum
Human Genome Variation (2018)