Abstract
Polydactyly is a clinically and genetically heterogeneous disorder. In the current report, we present a five-generation Chinese family with non-syndromic pre-axial polydactyly with thumb polydactyly (pre-axial polydactyly type I (PPD-I)) as a major clinical feature. Using whole-exome sequencing (WES), a novel nonsense mutation c.714T>A (p.Y238*) of the glioma-associated oncogene family zinc-finger 3 gene (GLI3) was identified as the pathogenic mutation for this family. Our study has, for the first time, suggested the possible contribution of GLI3 in the patheogenesis of PPD-I, and demonstrated that WES provided an applicable diagnostic tool for identifying mutations in disorders with highly genetical heterogeneity such as polydactyly.
Similar content being viewed by others
Introduction
Polydactyly is the most common digital malformation, which is characterized by whole or partial duplication of one or multiple digits.1 Digit duplication of polydactyly could be classified into two categories based on the location as follows: pre-axial (radial side) polydactyly with an extra thumb or hallux malformation, and post-axial (ulnar side) polydactyly with an extra little finger or toe.2, 3 Polydactyly is mostly inherited in an autosomal dominant manner,4 and over a 100 genes have been identified to be associated with human polydactyly.1, 5, 6, 7, 8, 9
In the current report, we undertook a clinical and genetic study of a Chinese family with non-syndromic bilateral pre-axial polydactyly (Supplementary Table 1) and identified a heterozygous nonsense mutation (c.714T>A /p.Y238*) of GLI3 gene as the putative disease-causing mutation. Such mutation was validated as co-segregated in this family by Sanger sequencing.
Our study demonstrated that WES provided a fast, accurate and cost-effective molecular diagnostic tool for identifying mutations in highly genetically heterogenous disorders, and broadened the phenotypic spectrum of GLI3 mutations.
Patients and methods
Members of a five-generation Chinese family with digit duplication of polydactyly were recruited with written consents, and the study was reviewed and approved by the ethics committee of Shanghai Children’s Medical Center.
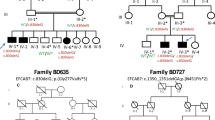
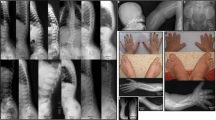
For all affected individuals in this family, the phenotypes were obtained on the basis of orthopedists’ diagnoses and the proband’s grandmother’s statements (Figure 1, Supplementary Table 1). A complete medical history was obtained, and a thorough physical examination was performed for all the participants in this study. Patients’ hands, feet and head were radiographed. Measurements included occipitofrontal (head) circumference, head shape, facial dysmorphism, brain imaging, inter-pupillary distance, inner-canthal distance, height or limb length, genitalia, anus and throat (epiglottis) were also examined in order to exclude other congenital malformations.
(a) Pedigree of a Chinese family with complex digital anomalies. Squares and circles denote males and females. Filled shapes indicate affected members. Individuals labeled with a solidus are deceased. An arrow denotes the proband. Members marked with * were participants in this study. (b) Clinical features of the proband (V-1). (c) Clinical features of the patient (III-2). (d) Clinical features of the patient (IV-2). A full color version of this figure is available at the Journal of Human Genetics journal online.
According to the Temtamy–McKusick classification,10 the major manifestation of the affected individuals in this family was pre-axial polydactyly type I (PPD-I; Supplementary Table 1). The propositus had bilateral PPD-I in both feet, and bilateral pre-axial polydactyly with incomplete syndactyly in both thumbs. The whole-exome sequencing (WES) was conducted to identify the pathogenic mutation(s), following experimental procedures and bioinformatics analysis pipelines described previously.11, 12 The findings of WES were validated by Sanger sequencing.
Results
The propositus was a 5-year-old child of a non-consanguineous parents, who had a symmetrical head with normal circumference (47.0 cm, normal range: 46.2–49.0 cm). The head circumference of all other family members studied was within normal range (adult male: 56.1–59.2 cm; adult female: 54.0–56.9 cm). The physiological and intellectual development of all family members in this study was also normal. Except thumb polydactyly, no other abnormality was observed in this family. The diagnosis of non-syndromic pre-axial polydactyly (PPD-I) was established based on the symptoms and physical examination, imaging studies and exclusion of differential diagnosis, such as Pallister–Hall syndrome, Greig Cephalopolysyndactyly syndrome and Acrocallosal syndrome.
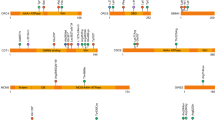
Among genetic variances detected by WES (Supplementary Table 2), there was a novel heterozygous mutation (c.714T>A, p.Y238*, Figure 2) in exon 6 of GLI3. Sanger sequencing was performed to validate the novel mutation in the family. As shown in Figure 2, this mutation co-segregated with the polydactyly phenotype in this family.
Novel single-nucleotide nonsense mutation detected in GLI3. (a) The structure of GLI3 and its product showing c.714T>A in exon 6, and the position of the resulting stop gain at amino acid 238. (b) Sanger sequencing of the PCR product at the mutated region. From top to bottom: human reference sequence, followed by the PCR sequence of three affected members (V-I, IV-2, III-2) with the same mutation c.714T>A. A full color version of this figure is available at the Journal of Human Genetics journal online.
Discussion
Polydactyly exhibits various clinical presentations and can be caused by mutations of over a 100 genes. The correlations between pathogenic genetic changes and polydactyly phenotypes have not been fully established. Therefore, it would be impractical to identify pathogenic mutation(s) using Sanger sequencing for a given case of polydactyly. The WES, due to its high efficiency, accurateness and cost-effectiveness in exome scanning, has become an ideal approach for patients with polydactyly to identify disease-causing mutations.
GLI3 protein has been shown to be associated with five distinct types of polydactyly disorders. Three of them are syndromes with polydactyly as a significant feature.9, 13, 14 Another two types are non-syndromic: post-axial polydactyly type A/B (PAP-A/B)15 and PPD-IV.16 In a previous work, three de novo mutations of GLI3 were identified in post-axial polydactyly cases, which were pathogenic given their loss of function impact on protein.17 In our present work, we identified a GLI3 mutation (p.Y238*) using WES analysis in a Chinese family with PPD-I, and to our knowledge, it is for the first time that GLI3 mutations has been linked to the pathogenesis of PPD-I. The genotype–phenotype correlation between the novel GLI3 mutation of c.714T>A (p.Y238*) and PPD-I phenotype expanded the phenotypic spectrum of GLI3 mutations.18, 19, 20
In conclusion, a novel GLI3 mutation c.714T>A (p.Y238*) was identified in a Chinese family with pre-axial polydactyly. Our results broadened the phenotypic spectrum of GLI3 mutations and demonstrated the feasibility of WES in clinical application of molecular diagnosis.
References
Biesecker, L. G. Polydactyly: how many disorders and how many genes? Am. J. Med. Genet. 112, 279–283 (2002).
Swanson, A. B. A classification for congenital limb malformations. J. Hand. Surg. 1, 8–22 (1976).
Malik, A. Polydactyly: phenotypes, genetics and classification. Clin. Genet. 85, 203–212 (2014).
Radhakrishna, U., Blouin, J. L., Mehenni, H., Patel, U. C., Patel, M. N., Solanki, J. V. et al. Mapping one form of autosomal dominant postaxial polydactyly type A to chromosome 7p15-q11.23 by linkage analysis. Am. J. Hum. Genet. 60, 597–604 (1997).
Wen-bin, H. E. & Wen, L. I. Finger (toe) abnormality and the latest relative genetic research. Progr. Mod. Med. 12, 5182–5188 (2012).
Debeer, P., Schoenmakers, E. F., Twal, W. O., Argraves, W. S., De Smet, L., Fryns, J. P. et al. The fibulin-1 gene (FBLN1) is disrupted in a t (12; 22) associated with a complex type of synpolydactyly. J. Med. Genet. 39, 98–104 (2002).
Park, K., Kang, J., Subedi, K. P., Ha, J. H. & Park, C. Canine polydactyly mutations with heterogeneous origin in the conserved intronic sequence of LMBR1. Genetics 179, 2163–2172 (2008).
Merrill, A. E., Merriman, B., Farrington-Rock, C., Camacho, N., Sebald, E. T., Funari, V. A. et al. Ciliary abnormalities due to defects in the retrograde transport protein DYNC2H1 in short-rib polydactyly syndrome. Am. J. Hum. Genet. 84, 542–549 (2009).
Vortkamp, A., Gessler, M. & Grzeschik, K. H. GLI3 zinc-finger gene interrupted by translocations in Greig syndrome families. Nature 352, 539–540 (1991).
Temtamy, S. A. & McKusick, V. A. The genetics of hand malformations. Birth Defects Orig Artic Ser. 14, i–xviii, 1–619 (1978).
Wang, J., Wang, Z. G., An, Y., Wu, C. X., Xu, Y. L., Fu, Q. H. et al. Exome sequencing reveals a novel PTHLH mutation in a Chinese pedigree with brachydactyly type E and short stature. Clin. Chim. Acta. 446, 9–14 (2015).
Wang, J., Zhang, W. X., Jiang, H. & Wu, B. L. Primary ovarian insufficiency collaboration. mutations in HFM1 in recessive primary ovarian insufficiency. N. Engl. J. Med. 370, 972–974 (2014).
Kang, S., Graham, J. M. Jr, Olney, A. H. & Biesecker, L. G. GLI3 frameshift mutations cause autosomal dominant Pallister-Hall syndrome. Nat. Genet. 15, 266–268 (1997).
Elson, E., Perveen, R., Donnai, D., Wall, S. & Black, G. C. De novo GLI3 mutation in Acrocallosal syndrome: broadening the phenotypic spectrum of GLI3 defects and overlap with murine models. J. Med. Genet. 39, 804–806 (2002).
Radhakrishna, U., Wild, A., Grzeschik, K. H. & Antonarakis, S. E. Mutation in GLI3 in postaxial polydactyly type A. Nat. Genet. 17, 269–271 (1997).
Radhakrishna, U., Bornholdt, D., Scott, H. S., Patel, U. C., Rossier, C., Engel, H. et al. The phenotypic spectrum of GLI3 morphopathies includes autosomal dominant preaxial polydactyly type-IV and postaxial polydactyly type-A/B; no phenotype prediction from the position of GLI3 mutations. Am. J. Hum. Genet. 65, 645–655 (1999).
Wang, Z. G., Wang, J., Li, Y. C., Geng, J., Fu, Q. H., Xu, Y. L. et al. Novel frame-shift mutations of GLI3 gene in non-syndromic postaxial polydactyly patients. Clin. Chim. Acta. 433, 195–199 (2014).
Démurger, F., Ichkou, A., Mougou-Zerelli, S., Le Merrer, M., Goudefroye, G., Delezoide, A. L. et al. New insights into genotype-phenotype correlation for GLI3 mutations. Eur. J. Hum. Genet. 23, 92–102 (2015).
Johnston, J. J., J Sapp, J. C., Turner, J. T., Amor, D., Aftimos, S., Aleck, K. A. et al. Molecular analysis expands the spectrum of phenotypes associated with GLI3 mutations. Hum. Mutat. 31, 1142–1154 (2010).
Biesecker, L. G. What you can learn from one gene: GLI3. J. Med. Genet. 43, 465–469 (2006).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Xiang, Y., Wang, Z., Bian, J. et al. Exome sequencing reveals a novel nonsense mutation of GLI3 in a Chinese family with ‘non-syndromic’ pre-axial polydactyly. J Hum Genet 61, 907–910 (2016). https://doi.org/10.1038/jhg.2016.76
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2016.76