Abstract
Many single-nucleotide polymorphisms (SNPs) in the non-recombining region of the human Y chromosome have been described in the last decade. High-coverage sequencing has helped to characterize new SNPs, which has in turn increased the level of detail in paternal phylogenies. However, these paternal lineages still provide insufficient information on population history and demography, especially for Native Americans. The present study aimed to identify informative paternal sublineages derived from the main founder lineage of the Americas—haplogroup Q-L54—in a sample of 1841 native South Americans. For this purpose, we used a Y-chromosomal genotyping multiplex platform and conventional genotyping methods to validate 34 new SNPs that were identified in the present study by sequencing, together with many Y-SNPs previously described in the literature. We updated the haplogroup Q phylogeny and identified two new Q-M3 and three new Q-L54*(xM3) sublineages defined by five informative SNPs, designated SA04, SA05, SA02, SA03 and SA29. Within the Q-M3, sublineage Q-SA04 was mostly found in individuals from ethnic groups belonging to the Tukanoan linguistic family in the northwest Amazon, whereas sublineage Q-SA05 was found in Peruvian and Bolivian Amazon ethnic groups. Within Q-L54*, the derived sublineages Q-SA03 and Q-SA02 were exclusively found among Coyaima individuals (Cariban linguistic family) from Colombia, while Q-SA29 was found only in Maxacali individuals (Jean linguistic family) from southeast Brazil. Furthermore, we validated the usefulness of several published SNPs among indigenous South Americans. This new Y chromosome haplogroup Q phylogeny offers an informative paternal genealogy to investigate the pre-Columbian history of South America.
Similar content being viewed by others
Introduction
The non-recombining region of the Y chromosome (NRY) is the subject of intense research in the field of human population genetics and evolution.1, 2, 3 Several studies of the peopling of the Americas have made use of Native American NRY polymorphic variants.4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22 The major founder lineage (or haplogroup) of Native American populations, Q-L54,4, 5, 6, 11, 16, 17 is defined by the derived allele of the L54 SNP and belongs to haplogroup Q, which in turn is defined by the derived allele at locus M242.12, 23, 24 Lineage Q-L54, which is currently divided in Native Americans into Q-L54*(xM3) and Q-M3, accounts for at least 85% of the autochthonous Y chromosomes of native Americans.12, 14, 15, 16, 17, 19, 20 A rare native American lineage is haplotype C-M217 (or C3),17 which is defined by marker M21721 and is derived from haplogroup C, which is in turn defined by markers M216 and M130.22 This latter haplogroup is also found among various ethnic groups from northeast Asia, Australasia and Oceania.25
The use of Y chromosome single-nucleotide plymorphisms (Y-SNPs) in human history reconstruction depends on their ability to discriminate between paternal lineages shared by individuals and populations.1, 10 Next-generation sequencing technologies have identified thousands of new SNPs, and these discoveries have significantly enhanced the resolution and topology of the Y-chromosomal phylogeny.3, 20, 26, 27, 28 However, many markers provide redundant information or define terminal phylogenetic branches that are not useful at the population level.24, 29
Genetic studies have traditionally combined Y-chromosomal SNPs and short tandem repeats (STRs). Hundreds of STRs on the human Y chromosome have been described,30 and their high variability allows phylogeographic studies of SNP-defined Y-chromosomal lineages shared by individuals from different ethnic groups.31, 32, 33, 34 Y-STR haplotype analyses have been used for the historical reconstruction of the peopling of Americas in population migration analyses, demographic estimates, dating of historical events and evaluations of paternal gene flow.12, 24, 33, 34, 35 However, because of the high mutation rate of Y-STRs,31 ancient SNP-defined lineages (for example, >10 000 years old) can present highly homoplasic phylogeographic networks, resulting in unresolved phylogenies with low genealogical information as depicted by network analyses of the Q-M3 lineage.17, 18 Thus, to increase the phylogeographic information of Y chromosome data, one can use SNPs defining more recent sublineages to generate a more detailed Y-STR genealogy, as previously shown.24, 31
Following this approach, we identified new informative Q-L54 sublineages among native South American populations by screening genealogically divergent haplogroup Q Y chromosomes, for 34 SNPs identified through re-sequencing work. They were genotyped in a large sample of South American natives together with SNPs available in the literature. Five new SNPs defining two Q-M3-derived and three Q-L54*(xM3)-derived sublineages were validated for use in phylogeographic studies of native South American populations. All informative SNPs were used to increase the resolution of the phylogeny of haplogroup Q lineages in combiantion with previously described SNPs, to build a more detailed and informative genealogy of native South American Y chromosomes.
Materials and methods
A general scheme of the procedures described below to identify and validate informative Y-SNPs for native South American population studies is depicted in Figure 1.
Sampling
The subjects of the study consisted of 1841 native American individuals (Supplementary Table 1) belonging to Y chromosome Q or C lineages (carrying the derived alleles at SNPs M242 or M130, respectively) from indigenous communities in Peru, Bolivia, Ecuador and Brazil, sampled during the Genographic Project,18, 24 and also 13 Coyaima individuals (Cariban linguistic family) from Colombia. DNA samples were obtained from cells collected via buccal swabs.18, 24 In the case of the Coyaima individuals, aliquots of DNA from a previous study24 were used. The present study was authorized in Brazil by the ethics commissions of the Universidade Federal de Minas Gerais and the National Commission for Ethical Research (CONEP, Resolution 763/2009), and by local ethics commissions of the non-Brazilian countries in which samples were collected.24
Initial genotyping of NRY SNPs and STRs
Native American samples were initially genotyped for NRY SNPs M242, M3 and M130 using custom TaqMan genotyping assays (Applied Biosystems) to identify haplogroup Q and C Y chromosomes (Supplementary Text 1). At a later stage, all samples were also genotyped in the BeadXpress system (see below) for the presence of the derived alleles at markers L54 and M217, which identify native American lineages Q-L54 and C-M217, respectively. The samples were also genotyped using 17 Y-STRs (DYS389a, DYS389b, DYS390, DYS456, DYS19, DYS385a, DYS385b, DYS458, DYS437, DYS438, DYS448, GATA_H4, DYS391, DYS392, DYS393, DYS439, DYS635) with a Y-filer Kit x(Applied Biosystems, Foster City, CA, USA) following manufacturer’s recommended protocol36 (Supplementary Text 1).
Sample selection for large-scale sequencing
Using the median-joining method implemented in NETWORK 4.6,37 haplotype networks were constructed with Y-STR data from South American individuals belonging to the Q-M3 and Q-L54* lineages, as described by Jota et al.24 Samples submitted for Y chromosome sequencing were selected by choosing Y-STR haplotypes located in separated network clusters, giving priority to representatives of different ethnicities and geographic regions from South America. This selection based on Y-STRs was used to increase the chance to identify new SNPs defining sublineages on different branches of Q-M3 and Q-L54* lineages.
Identification of new Y-SNPs in haplogroup Q
To identify new Y-SNPs, NRY regions from 21 samples of indigenous South Americans belonging to the haplogroup Q-L54 were sequenced using different strategies. Sixteen individuals were analyzed via Sanger sequencing, two individuals via long-PCR and next-generation sequencing, two individuals via the Walk Through the Y (WTY) sequencing service (Family Tree DNA), and one individual via complete Y chromosome sequencing (Complete Genomics). The individuals were selected using the previously defined phylogenetic, geographic and ethnic criteria.
Among the 16 samples selected for Sanger sequencing (two Bolivian, six Brazilian, two Colombian and six Peruvian Indians), six belonged to the Q-L54*(xM3) paragroup and 10 to the Q-M3 lineage. The samples were subjected to de novo sequencing after PCR amplification of known Y chromosome regions at loci M323,38 M378,39 MEH2,1 N14,40 P292,10 M242,23 M3,41 M138, M217,42 Page53,12, 13 Alu486, DFFRY04, DBYd1 and P8943 (Supplementary Text 1).
High-coverage next-generation sequencing from long-PCR-amplified fragments was performed using two samples from the Q-M3 lineage (1 each from Peru and Brazil). This sequencing involved the generation of 672 DNA segments via long PCR using Taq HiFi (Invitrogen), covering about 3 million base pairs of the NRY region3, 44 (Supplementary Text 1).
Two samples from native Peruvians belonging to the Q-M3 lineage underwent Sanger sequencing using the WTY (Walk Through the Y) service at the Genomic Research Center of Family Tree DNA.45 This protocol generates sequences over 180 000 bp of the NRY region and returns a list of probable SNPs identified from single copy, euchromatic regions of the human Y chromosome.
One sample from the Q-M3 lineage originating from Peru was also sent for complete genome sequencing at Complete Genomics.46
Y-SNP multiplex genotyping
Two 96 Y-SNPs platforms were constructed by designing a personalized multiplex genotyping system using the VeraCode OPA system (Illumina, San Diego, CA, USA, Supplementary Text 1) and performing genotype reading using the BeadXpress system (Supplementary Tables 2 and 3). Allowing for duplications of SNPs included in both platforms, a total of 119 SNPs were tested (Table 2). We evaluated 34 new SNPs (Table 1) that were identified in our study, and 85 previously known Y chromosome SNPs: 28 SNPs identifying haplogroup Q sublineages, including L54;10, 45, 47 22 SNPs that were previously identified in various studies through Y chromosome sequencing of native Americans, but were not evaluated at the population level in South America;3, 47, 48 and 35 SNPs identifying other Y haplogroups.1, 10 The 119 Y-SNPs, and their chromosomal positions, which were used on the two multiplex genotyping platforms are listed in Table 2.
The two multiplex sets of 96 SNPs were referred to as platform 1 (VC0014123-OPA) and platform 2 (VC0014259-OPA). Platform 2 was constructed in a second stage, with the inclusion of SNPs that were validated on platform 1 and shown to be shared among native South Americans. Details of the BeadXpress genotyping assay were published elsewhere. 49, 50
Native South Americans were submitted to multiplex genotyping using BeadXpress platforms 1 and 2, in a hierarchical procedure defined by the Y chromosome tree1, 10 and a screening method based on the Y-STR profile (see below). The BeadXpress genotyping assays for SNPs SA02 and SA03 were not successful, thus they were genotyped by standard restriction fragmentation length polymorphism (RFLP) analysis (Supplementary Text 1). In total, 960 samples (including also controls, data not shown) were fully genotyped using both platforms (Supplementary Tables 2 and 3).
All new SNPs were incorporated into the Y-chromosomal tree according to the haplogroup hierarchy and nomenclature defined by the Y Chromosome Consortium.1, 10
Screening of samples for NRY SNP genotyping
Using the Y-STR haplotype networks from South American individuals belonging to the Q-M3 and Q-L54* lineages, samples were selected for genotyping new NRY SNPs on BeadXpress or PCR–RFLP genotyping (SA02 and SA03 SNPs). Samples were selected by choosing Y-STR haplotypes phylogenetically related to haplotypes carrying new SNPs (Supplementary Figure 1). This process was repeated when new individuals bearing new Y-SNPs were found. In addition, all individuals from a location or ethnic group where a new SNP was found were also selected for genotyping each particular SNP.
Dating of new South American Q-L54 sublineages
Estimates of the time to the most recent common ancestor (TMRCA) for chromosomes carrying the derived alleles of the new lineage-defining SNP stemming from the present study were determined using rho statistics, implemented in the NETWORK program,37 employing a mean effective mutation rate of 6.9 × 10−4/locus/25 years51 for each Y-STR locus. The ancestral haplotype was inferred using the modal allele at each STR locus.32
Results
Our initial survey of Y chromosome variation in South American natives with TaqMan RT-PCR identified 1,836 Q-M242 and five C-M130 haplogroup individuals. Because there is an overwhelming predominance of haplogroup Q chromosomes in South America, we focused on SNP identification and validation of new Q-derived lineages. Using four different sequencing strategies for the NRY region, we identified a total of 2503 putatively new Y-chromosomal SNPs. All Y-SNPs were aligned according to their relative position against a reference genome (hg19/GRCh37) and were selected for validation via multiplex genotyping in a BeadXpress system (Illumina). The SNP selection criteria for inclusion in the genotyping platforms were (i) previously unknown variable positions in high-quality sequenced regions, particularly transversions, (ii) SNPs shared between two or more individuals, and (iii) SNPs from individuals representing different ethnic and geographic backgrounds.
In the present study, 34 Y-SNPs were initially selected from the different sequencing strategies to generate BeadXpress genotyping platforms 1 and 2 (Table 1). The positions of the SNPs and the sequencing methodology used to identify them are summarized in Table 1. Altogether with the new Y-SNPs found here, we included most of the SNPs described in the literature for haplogroup Q,1, 10, 12, 16, 17, 20, 24, 29 particularly the ones likely informative for discriminating among South American chromosomes, such as L53, L54, SA01, Z5915, Z19483, Z19319, CTS11357, M19 and CTS1780 (Table 2). The M557 and PV2 SNPs16 were not tested in our surveys.
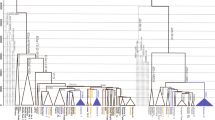
Using multiplex genotyping platforms 1 and 2, we identified three new informative SNPs in the Q-L54 lineage—SA04, SA05 and SA29—which were validated for studies at the population level. Two other population-informative SNPs, SA02 and SA03 (SA03.2), were genotyped by Sanger sequencing and RFLP analysis because the BeadXpress assay failed for these two markers. Furthermore, we validated through the BeadXpress genotyping system an additional five SNPs—Z5915, Z19483, Z19319;47 CTS11357;3 and CTS1780 (ref. 48; Supplementary Tables 2 and 3). Many synapomorphic SNPs, shared by different individuals, are shown in the new Q-haplogroup phylogeny based on our results (Figure 2). For each of these new Q-L54 sublineages (except Q-SA29), we estimated the TMRCA for the chromosomes carrying the derived allele (Table 3).
Haplogroup Q Phylogeny. Phylogeny of Q haplogroup based on Y-SNPs genotyped (Table 2) in this study. SNP, single-nucleotide polymorphism; Y-SNPs, Y chromosome SNPs A full color version of this figure is available at the Journal of Human Genetics journal online.
The SA04 SNP consists of an A->T transversion at Y position 15974563 (GRCh37/hg19) and is found in haplogroup Q-M3. We identified 45 individuals exhibiting the SA04-derived allele, 37 of whom were from five Brazilian indigenous communities (from the municipality of São Gabriel da Cachoeira, Amazonas State, northwest Amazon of Brazil), seven from Ecuador, and one from Peru (Supplementary Tables 4 and 5). We tested a total of 667 haplogroup Q-M3 samples for SA04 using multiplex platforms 1 and 2 (Table 2). Among the 45 individuals displaying the SA04-derived allele, seven Andeans were from Ecuador, one individual belonged to the Muniche community (language isolate) from Loreto (Peru), and 37 individuals were from the cultural confluence area of the northwest Brazilian Amazon on the Upper Negro River (Supplementary Tables 4 and 5). A phylogeographic network was constructed with the Y-STR haplotypes of SA04-derived allele-carrying individuals with their linguistic families (Supplementary Figure 2). The geographic and cultural connectedness of the Q-SA04 lineage individuals corroborates the hypothesis of its recent genealogical origin.
The SA05 SNP is an A->G transition at Y position 8148836 (GRCh37/hg19) found in haplogroup Q-M3. We identified 60 Amazonian individuals exhibiting the SA05-derived allele, 40 of whom originated from 13 different Peruvian indigenous communities, and 20 from 6 indigenous communities from Bolivia (Supplementary Tables 4 and 5). We tested a total of 667 haplogroup Q-M3 samples for SA05 using SNP multiplex platforms 1 and 2 (Table 2).
The SA29 SNP consists of a C->T transition at Y position 6931891 (GRCh37/hg19) found in lineage Q-L54*(xM3). We identified five individuals carrying the SA29 derived allele (Supplementary Tables 4 and 5) from the Maxacali indigenous community, which belongs to the Jean linguistic family, from Minas Gerais (Brazil). We tested a total of 117 Q-L54* individuals for SA29 using SNP multiplex platform 2 (Table 2). Even though these Q-SA29 individuals were apparently unrelated (at first degree) and from different locations, they have the same Y-STR haplotype (Supplementary Table 4).
The SA02 and SA03 (SA03.2) SNPs were tested with multiplex genotyping platforms 1 and 2, with unsatisfactory results. Thus, new genotyping assays based on RFLP analyses were developed for both SNPs (Supplementary Text 1). SA03 actually includes two linked SNPs (SA03.1 and SA03.2) at the DBYd1 locus, which detect the same lineage (Supplementary Text 1). SA03.2 consists of an A->G transition at position 15019822 (GRCh37/hg19), and SNP SA03.1 consists of a C->G transversion at position 15019808 (GRCh37/hg19), which is separated from SA03.2 by 13 base pairs. Both the SA03.1 and the SA03.2 SNPs were detected in eight Coyaima individuals (Cariban linguistic family) by sequencing and RFLP analysis of PCR products amplified using the DBYd1 Alu region primers (Supplementary Text 1). The SA02 SNP consists of an A->G transition at Y chromosome position 14820439 (GRCh37/hg19) in the DFFRY04 locus (Supplementary Text 1). We identified seven individuals exhibiting the SA02 derived allele in the Coyaima population (Cariban linguistic family) from Colombia (Supplementary Tables 4 and 5). All seven individuals with the SA02 derived allele also exhibit the SA03.1 and SA03.2 derived alleles, but a single Coyaima individual carried the derived SA03.1 and SA03.2 alleles and the ancestral SA02 allele. The eight individuals tested for the three aforementioned SNPs also carried derived alleles for markers M242 and L54 and the ancestral allele for marker M3. In the sample set tested in the present study, SA02 and SA03 (SA03.1 and SA03.2) occur exclusively in the Coyaima population of Colombia.
The BeadXpress multiplex platform 2 (Table 2) was also used to characterize the previously described Q sublineages defined by SNPs CTS11357, CTS1780, M19, Z19483, Z19319 and Z5915 (Figure 2). We tested a total of 272 Q-M3 samples for CTS11357 using multiplex platform 2, and identified eight individuals from Western Amazon exhibiting the derived alleles, seven of whom originated from three different Peruvian indigenous communities, while one originated from Bolivia (Supplementary Tables 4 and 5).
We tested a total of 660 haplogroup Q-M3 samples for the M19 SNP using multiplex platforms 1 and 2 (Table 2). We tested a total of 272 Q-M3 haplogroup samples for the Z5915 SNP using multiplex platform 2 (Table 2), and identified two individuals with the derived allele for Z5951 in haplogroup Q-M3 (Supplementary Tables 4 and 5). We tested a total of 117 Q-L54* samples for the CTS1780 SNP using multiplex platform 2 (Table 2), and identified 112 individuals exhibiting the derived state. Although five individuals failed to genotype in BeadXpress, all genotyped South American Q-L54* chromosomes were included in a new derived lineage, Q-CTS1780. This lineage is widely distributed in Peru, Bolivia and Ecuador (Supplementary Table 4 and 5), and found among eight Coyaima individuals from Colombia also bearing the SA03 SNP, and five individuals from the Maxacali community (Jean linguistic family) from Minas Gerais (Brazil), who also had the SA29 derived allele. Both Q-SA29 and Q-SA03 are sublineages within Q-CTS1780 (Figure 2). The TMRCA (Table 3) of Q-CTS1780 reveals it to be an ancient lineage, likely predating the first entry into Americas by the first settlers.4, 16, 17
We initially tested a total of 231 Q-M3 individuals for the Z19319 SNP using multiplex platform 2 (Table 2), and identified 21 haplogroup Q-M3 individuals exhibiting the derived allele. Twenty Q-Z19319 individuals were from Andean indigenous communities of Peru and Bolivia (Supplementary Table 4 and 5). Thirteen individuals exhibiting the Z19319 derived allele also carried the SA0124 derived allele (Supplementary Tables 4 and 5). Q-SA01 now constitutes a sublineage within Q-Z19319 (Figure 2). The eight individuals with the Z19319 derived allele and the SA01 ancestral allele are now classified in the Q-Z19319*(xSA01) paragroup (Figure 2). One individual of paragroup Q-Z19319* is found in each location, except for Chogo (northern Central Andes), which presented two of them (Supplementary Tables 4 and 5). The TMRCA for the Q-Z19319 lineage (Table 3) indicated an origin in the Holocene (~9,000 years ago), and likely in the northern region of Central Andes (Cajamarca, Peru), as it is also true for its derivative lineage, Q-SA01, which was dated at about 5,300 years ago.24
We tested a total of 323 Q-M3 haplogroup samples for the Z19483 SNP using multiplex platform 2 (Table 2), and identified 60 haplogroup Q-M3 individuals carrying the Z19483-derived allele. Most of them were Andeans, with 41 individuals coming from 21 different Bolivian indigenous communities, and 19 from 12 different Peruvian indigenous communities (Supplementary Tables 4 and 5).
We also identified a single Q-L53* individual from Quinuabamba, Peru, among our total sample (Supplementary Tables 4 and 5). Even though we identified a large set of new Q-M3 sublineages, Q-M3* paragroup individuals are still the most frequent and widely distributed chromosomes, being observed in 462 individuals who were genotyped using multiplex platforms 1 and 2 (Supplementary Tables 2 and 3).
The five haplogroup C (C-M130) individuals originated from two Ecuadorian communities, and one from Peru. All five individuals were Quechua speakers (Supplementary Table 4), and displayed also the derived allele for SNP M217; thus, these South American natives belong to the C-M217 lineage, as previously reported.17
Supplementary Tables 2 and 3 summarize the Y-SNP genotyping results. Of the 960 samples, 172 served as controls for haplogroups or duplicates, and 10 failed to be genotyped using BeadXpress.
Discussion
The use of the BeadXpress multiplex genotyping platform contributed to the proposal of a new haplogroup Q phylogeny (Figure 2), which differs from those proposed by Van Oven et al.29 and Geppert et al.20 in its enhanced resolution to discriminate between South American indigenous lineages. Within the Q-L54 lineage, South American individuals were divided into the Q-CTS1780 and Q-M3 sublineages, which represent two ancient lineages (>20 000 years old) that likely arrived concomitantly or split early during the first peopling of Americas. These results are in partial agreement with previous reports.11, 14, 15, 16, 17, 18, 20
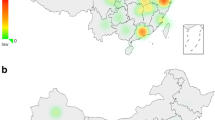
A significant difference in the phylogeny proposed in the present study is related to the Q-SA01 lineage, which is now derived from the Q-Z19319 lineage. Considering the distribution of SA01 chromosomes shown in the map of Figure 3, and the occurrence of Q-Z19319*(xSA01) chromosomes in a more northerly part of the Central Andes in Peru, our findings corroborate an Andean migratory route from north to south as previously suggested by the analysis of Y-STRs of the Q-SA01 lineage.24
Distribution of derived alleles for new Y-SNPs found in this study among native South Americans. A map of South America showing locations of indigenous communities displaying haplogroup Q Y chromosomes with derived alleles at SA01, SA03, SA04, SA05, and SA29 SNPs. Individuals displaying the derived SA02 allele are part of the Q-SA03 lineage. SNP, single-nucleotide polymorphism; Y-SNPs, Y chromosome SNPs.
All new lineages found in this study have a restricted spatial distribution (Figure 3), showing that the inclusion of new SNPs also increases the degree of geographical association, which was weakly observed when only Y-STRs within the entire Q-M3 lineage were considered.17 For example, the SA05 SNP is found scattered in lower altitude regions of the pre-Andes Amazon from Peru and Bolivia, while the SA04 SNP is found in regions of the northwestern Amazon and northern Central Andes (Ecuador), displaying a strong association with indigenous populations of the Tukanoan linguistic family. The findings of many unrelated and younger branches in the haplogroup Q phylogeny (Figure 2) is expected considering the population expansions occurring during the settlement of Americas, including more recent local and regional expansions as already shown for native American Y chromosome data by Battaglia et al. 16
The sublineage Q-SA05 occurs exclusively in western Amazon communities of Bolivia and Peru, some living in transition rainforest areas (Yunga) between lowland Amazon and the Andes. Interestingly, it was found in 60 individuals speaking 15 Amazonian languages belonging to five linguistic families, and also two language isolates. Because SA05 appears to have a relatively old origin (TMRCA ~10 000 years ago, Table 3), it was expected to have a broad distribution throughout indigenous communities speaking different languages. However, owing to its restricted geographic distribution and occurrence in many indigenous populations, it could alternatively be an ancient lineage belonging to local hunter-gatherers who were assimilated into modern groups. This restricted distribution could also be attributed to genetic drift, which is much more pronounced among Amazonian indigenous groups than Andean ones.33
Individuals belonging to the Q-SA04 lineage are restricted to the northwestern Amazon, across Brazil, Peru and Ecuador. We identified the probable root of the Q-SA04 lineage as the ancestral Y-STR haplotype (IAU06) based on each STR locus’ modal allele (Supplementary Table 4). The Y-STR data collected in the present study suggest a likely migratory route for lineage Q-SA04 from the Brazilian Amazon to the Andes, as the haplotypes carrying the modal alleles were located in Iauaretê, on the border between Brazil and Colombia. The presence of more recent and derived Q-SA04 STR haplotypes in the Andes (Ecuador) may indicate that Amazonian groups were assimilated into the Central Andes indigenous communities (Quechua and Aymara) as previously suggested,18 likely during the formation of the Andean states, which culminated with the Inca Empire. However, genotyping a larger number of samples from this region will allow further clarification of this matter.
Interestingly, the derived allele of the SA04 SNP appears in 37 of the 63 individuals from São Gabriel da Cachoeira, in the Upper Rio Negro (northwestern Brazilian Amazon, close to the border with Colombia). Twenty-nine of them belong to the Tukanoan linguistic family, six to the Arawakan linguistic family and two to the Puinavean linguistic family. The Tukano and Arawak groups are horticulturalist communities who interact culturally in the region through the exchange of wives. By contrast, the Puinavean people are typical endogamous hunter–gatherer groups, but sometimes allow marriages between Tukano males and Puinavean females.52
The SA03 and SA02 SNPs were found in a single ethnic group sampled from Colombia, although those chromosomes display different Y-STR haplotypes and a TMRCA of about 1000 years (Table 3). Therefore, a larger population survey in Colombia and neighboring regions is needed to reveal their true distribution among native South Americans. The SA29 SNP is the first Y-SNP described for populations from the Jean linguistic group. However, even though a careful sampling was done to avoid relatives up to third degree, all the individuals bearing SA29 also have the same Y-STR haplotype. Thus, it is likely that this SNP has a very recent origin, although a larger population survey is still needed to confirm this hypothesis.
The distribution of Y-SNPs also provides clues about transition areas for the occurrence of paternal lineages. The area between Puerto Ocopa and Mazamari, in Peru, which is a boundary region between the Andes mountain range and the Amazon plain, exhibits the greatest haplotypic diversity among Q chromosomes, with five different lineages being observed there, including Q-CTS11357 (n=2); Q-Z19319* (n=2); Q-SA01 (n=1); Q-SA05 (n=9); Q-M3* (n=6; Supplementary Table 5). This is also the area where the greatest number of Andean (Q-Z19319* and SA01) and lowland (Q-SA05 and Q-CTS11357) lineages were observed, indicating a point of contact between Andean and Amazon populations (Figure 3). Indeed, it is located in an Andes-Amazon transition region of the Junin Province (Peru) where many natives from Andean (Quechua) and Amazonian (Ashaninka, Nomatsiguenga, Kakinte and Yanesha) origins (INEI, Peru) live.
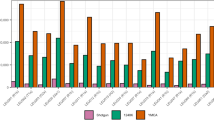
The TMRCA estimates (Table 3) for each Q sublineage based on Y-STR variation also give clues about their association with particular cultural shifts and demographic events, such as the suggested association of maize cultivation spread in the Andes with Q-SA01.24 According to Scliar et al.,53 lineages Q-SA05 (Amazonian) and Q-Z19319 (Andean) found in Peru are related to Pre-Ceramic periods I and II, when the cultivation of cassava, pumpkin and sweet potato was starting. In addition, lineage Q-CTS11357 in Peru may be related to Pre-Ceramic periods III and IV in the Amazon region, without showing a link to the cultivation of any particular vegetable. Lineage Q-SA01 is related to Pre-Ceramic periods V and VI and cultivation of many plants, including maize. Lineage Q-Z19483 is related to the Late Intermediate Period, marked by the founding of the Inca civilization, and could be an important marker for the study of the formation of the Andean states in the last millennium. Interestingly, Q-Z19483 is the most recent lineage (Table 3), but occurs in 60 native individuals distributed throughout Central Andes, which could be a likely outcome of a recent population expansion associated with a complex civilization like the Incas. However, these date estimates based on Y-STR and distance-based statistics should be interpreted with care because a number of issues have been raised.54
The discovery of new SNPs that are useful from the population point of view is important for acquiring further knowledge about the history of the peopling of South America. We were able to allocate approximately 30% of the Q-M3 lineage samples into derived sublineages. The new proposed phylogeny enhances the resolution of haplogroup Q in native Americans by including new Y-SNPs and connecting branches, particularly for populations in the Western Amazon and Andes.
Change history
31 March 2016
This article has been corrected since Advance Online Publication, and an erratum is also printed in this issue.
References
Y Chromosome Consortium A nomenclature system for the tree of human Y-chromosomal binary haplogroups. Genome Res. 12, 339–348 (2002).
Skaletsky, H., Kuroda-Kawaguchi, T., Minx, P. J., Cordum, H. S., Hillier, L., Brown, L. G. et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 423, 825–837 (2003).
Wei, W., Ayub, Q., Chen, Y., McCarthy, S., Hou, Y., Carbone, I. et al. A calibrated human Y-chromosomal phylogeny based on resequencing. Genome Res. 23, 388–395 (2013).
Pena, S. D., Santos, F. R., Bianchi, N. O., Bravi, C. M., Carnese, F. R., Rothhammer, F. et al. A major founder Y-chromosome haplotype in Amerindians. Nat. Genet. 11, 15–16 (1995).
Santos, F. R., Hurtz, M. H., Coimbra, C. E. A., Santos, R. V., Salzano, F. M. & Pena, S. D. Further evidence of existence of major founder Y chromosome haplotype in Amerindians. Braz. J. Genet. 18, 669–672 (1995).
Santos, F. R., Rodriguez-Delfin, L., Pena, S. D., Moore, J. & Weiss, K. M. North and South Amerindians may have the same major founder Y chromosome haplotype. Am. J. Hum. Genet. 58, 1369–1370 (1996).
Santos, F. R., Pandya, A., Tyler-Smith, C., Pena, S. D., Schanfield, M., Leonard, W. R. et al. The central Siberian origin for native American Y chromosomes. Am. J. Hum. Genet. 64, 619–628 (1999).
Underhill, P. A., Jin, L., Lin, A. A., Mehdi, S. Q., Jenkins, T., Vollrath, D. et al. Detection of numerous Y chromosome biallelic polymorphisms by denaturing high-performance liquid chromatography. Genome Res. 7, 996–1005 (1997).
Karafet, T. M., Zegura, S. L., Posukh, O., Osipova, L., Bergen, A., Long, J. et al. Ancestral Asian source(s) of new world Y-chromosome founder haplotypes. Am. J. Hum. Genet. 64, 817–831 (1999).
Karafet, T. M., Mendez, F. L., Meilerman, M. B., Underhill, P. A., Zegura, S. L. & Hammer, M. F. New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree. Genome Res. 18, 830–838 (2008).
Zegura, S. L., Karafet, T. M., Zhivotovsky, L. A. & Hammer, M. F. High-resolution SNPs and microsatellite haplotypes point to a single, recent entry of native American Y chromosomes into the Americas. Mol. Biol. Evol. 21, 164–175 (2004).
Dulik, M. C., Owings, A. C., Gaieski, J. B., Vilar, M. G., Andre, A., Lennie, C. et al. Y-chromosome analysis reveals genetic divergence and new founding native lineages in Athapaskan- and Eskimoan-speaking populations. Proc. Natl Acad. Sci. USA 109, 8471–8476 (2012a).
Dulik, M. C., Zhadanov, S. I., Osipova, L. P., Askapuli, A., Gau, L., Gokcumen, O. et al. Mitochondrial DNA and Y chromosome variation provides evidence for a recent common ancestry between native Americans and Indigenous Altaians. Am. J. Hum. Genet. 90, 229–246 (2012b).
Bisso-Machado, R., Jota, M. S., Ramallo, V., Paixão-Côrtes, V. R., Lacerda, D. R., Salzano, F. M. et al. Distribution of Y-chromosome Q lineages in native Americans. Am. J. Hum. Biol. 23, 563–566 (2011).
Bisso-Machado, R., Bortolini, M. C. & Salzano, F. M. Uniparental genetic markers in South Amerindians. Genet. Mol. Biol. 35, 365–387 (2012).
Battaglia, V., Grugni, V., Perego, U. A., Angerhofer, N., Gomez-Palmieri, J. E., Woodward, S. R. et al. The first peopling of South America: new evidence from Y-chromosome haplogroup Q. PLoS ONE 8, e71390 (2013).
Roewer, L., Nothnagel, M., Gusmão, L., Gomes, V., González, M., Corach, D. et al. Continent-wide decoupling of Y-chromosomal genetic variation from language and geography in native South Americans. PLoS Genet. 9, e1003460 (2013).
Sandoval, J. R., Lacerda, D. R., Jota, M. S., Salazar-Granara, A., Vieira, P. P., Acosta, O. et al. The genetic history of indigenous populations of the Peruvian and Bolivian Altiplano: the legacy of the Uros. PLoS One 8, e73006 (2013).
Geppert, M., Baeta, M., Núñez, C., Martínez-Jarreta, B., Zweynert, S., Cruz, O. W. et al. Hierarchical Y-SNP assay to study the hidden diversity and phylogenetic relationship of native populations in South America. Forensic Sci. Int. Genet. 5, 100–104 (2011).
Geppert, M., Ayub, Q., Xue, Y., Santos, S., Ribeiro-Dos-Santos, A., Baeta, M. et al. Identification of new SNPs in native South American populations by resequencing the Y chromosome. Forensic Sci. Int. Genet. 15, 111–114 (2015).
Mezzavilla, M., Geppert, M., Tyler-Smith, C., Roewer, L. & Xue, Y. Insights into the origin of rare haplogroup C3* Y chromosomes in South America from high-density autosomal SNP genotyping. Forensic Sci. Int. Genet. 15, 115–120 (2015).
Hallast, P., Batini, C., Zadik, D., Maisano Delser, P., Wetton, J. H., Arroyo-Pardo, E. et al. The Y-chromosome tree bursts into leaf: 13,000 high-confidence SNPs covering the majority of known clades. Mol. Biol. Evol. 32, 661–673 (2015).
Seielstad, M., Yuldasheva, N., Singh, N., Underhill, P., Oefner, P., Shen, P. et al. A novel Y-chromosome variant puts an upper limit on the timing of first entry into the Americas. Am. J. Hum. Genet. 73, 700–705 (2003).
Jota, M. S., Lacerda, D. R., Sandoval, J. R., Vieira, P. P., Santos-Lopes, S. S., Bisso-Machado, R. et al. A new subhaplogroup of native American Y-Chromosomes from the Andes. Am. J. Phys. Anthropol. 146, 553–559 (2011).
Zhong, H., Shi, H., Qi, X. B., Xiao, C. J., Jin, L., Ma, R. Z. et al. Global distribution of Y-chromosome haplogroup C reveals the prehistoric migration routes of African exodus and early settlement in East Asia. J. Hum. Genet. 55, 428–435 (2010).
Drmanac, R., Sparks, A. B., Callow, M. J., Halpern, A. L., Burns, N. L., Kermani, B. G. et al. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science 327, 78–81 (2010).
Poznik, G. D., Henn, B. M., Yee, M. C., Sliwerska, E., Euskirchen, G. M., Lin, A. A. et al. Sequencing Y chromosomes resolves discrepancy in time to common ancestor of males versus females. Science 341, 562–565 (2013).
Scozzari, R., Massaia, A., Trombetta, B., Bellusci, G., Myres, N. M., Novelletto, A. et al. An unbiased resource of novel SNP markers provides a new chronology for the human Y chromosome and reveals a deep phylogenetic structure in Africa. Genome Res. 24, 535–544 (2014).
Van Oven, M., Van Geystelen, A., Kayser, M., Decorte, R. & Larmuseau, M. H. Seeing the wood for the trees: a minimal reference phylogeny for the human Y chromosome. Hum. Mutat. 35, 187–191 (2014).
Gusmão, L., Butler, J. M., Carracedo, A., Gill, P., Kayser, M., Mayr, W. R. et al. DNA Commission of the International Society of Forensic Genetics (ISFG): an update of the recommendations on the use of Y-STRs in forensic analysis. Forensic Sci. Int. 157, 187–197 (2006).
Santos, F. R. & Tyler-Smith, C. Reading the human Y chromosome: the emerging DNA markers and human genetic history. Braz. J. Genet. 19, 665–670 (1996).
Zerjal, T., Dashnyam, B., Pandya, A., Kayser, M., Roewer, L., Santos, F. R. et al. Genetic relationships of Asians and Northern Europeans, revealed by Y-chromosomal DNA analysis. Am. J. Hum. Genet. 60, 1174–1183 (1997).
Tarazona-Santos, E., Carvalho-Silva, D. R., Pettener, D., Luiselli, D., De Stefano, G. F., Labarga, C. M. et al. Genetic differentiation in South Amerindians is related to environmental and cultural diversity: evidence from the Y chromosome. Am. J. Hum. Genet. 68, 1485–1496 (2001).
Bortolini, M. C., Salzano, F. M., Thomas, M. G., Stuart, S., Nasanen, S. P., Bau, C. H. et al. Y-chromosome evidence for differing ancient demographic histories in the Americas. Am. J. Hum. Genet. 73, 524–539 (2003).
Bailliet, G., Ramallo, V., Muzzio, M., García, A., Santos, M. R., Alfaro, E. L. et al. Brief communication: Restricted geographic distribution for Y-Q* paragroup in South America. Am. J. Phys. Anthropol. 140, 578–582 (2009).
Mulero, J. J., Chang, C. W., Calandro, L. M., Green, R. L., Li, Y., Johnson, C. L. et al. Development and validation of the AmpFlSTR Yfiler PCR amplification kit: a male specific, single amplification 17 Y-STR multiplex system. J. Forensic Sci. 51, 64–75 (2006).
Bandelt, H. J., Forster, P. & Röhl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16, 37–48 (1999).
Shen, P., Lavi, T., Kivisild, T., Chou, V., Sengun, D., Gefel, D. et al. Reconstruction of patrilineages and matrilineages of Samaritans and other Israeli populations from Y-chromosome and mitochondrial DNA sequence variation. Hum. Mutat. 24, 248–260 (2004).
Sengupta, S., Zhivotovsky, L. A., King, R., Mehdi, S. Q., Edmonds, C. A., Chow, C. E. et al. Polarity and temporality of high-resolution y-chromosome distributions in India identify both indigenous and exogenous expansions and reveal minor genetic influence of Central Asian pastoralists. Am. J. Hum. Genet. 78, 202–221 (2006).
Deng, W., Shi, B., He, X., Zhang, Z., Xu, J., Li, B. et al. Evolution and migration history of the Chinese population inferred from Chinese Y-chromosome evidence. J. Hum. Genet. 49, 339–348 (2004).
Underhill, P. A., Jin, L., Zemans, R., Oefner, P. J. & Cavalli-Sforza, L. L. A pre-Columbian Y chromosome-specific transition and its implications for human evolutionary history. Proc. Natl Acad. Sci. USA 93, 196–200 (1996).
Underhill, P. A., Passarino, G., Lin, A. A., Shen, P., Mirazón Lahr, M., Foley, R. A. et al. The phylogeography of Y chromosome binary haplotypes and the origins of modern human populations. Ann. Hum. Genet. 65, 43–62 (2001).
Wilder, J. A., Kingan, S. B., Mobasher, Z., Pilkington, M. M. & Hammer, M. F. Global patterns of human mitochondrial DNA and Y-chromosome structure are not influenced by higher migration rates of females versus males. Nat. Genet. 36, 1122–1125 (2004).
Ayub, Q., Jostins, L., Xue, Y., Turner, D. & Tyler-Smith, C. The 1000 Genomes Consortium Next generation sequencing and the era of personal Y genomes. Genome Biol. 11 (Suppl 1), O2 (2010).
Thomas Krahn/FTDNA (2001) https://www.familytreedna.com. Accessed 6 December 2012.
Complete Genomics (2013) http://www.completegenomics.com. Accessed 16 January 2013.
International Society of Genetic Genealogy. ISOGG Y-DNA Haplogroup Tree 2011, Version 9.129. (2011) http://isogg.org/tree. Accessed 4 December 2012.
Elhaik, E., Greenspan, E., Staats, S., Krahn, T., Tyler-Smith, C., Xue, Y. et al. The GenoChip: a new tool for genetic anthropology. Genome Biol. Evol. 5, 1021–1031 (2013).
Shen, R., Fan, J. B., Campbell, D., Chang, W., Chen, J., Doucet, D. et al. High-throughput SNP genotyping on universal bead arrays. Mutat. Res. 573, 70–82 (2005).
Lin, C. H., Yeakley, J. M., McDaniel, T. K. & Shen, R. Medium- to high-throughput SNP genotyping using VeraCode microbeads. Methods Mol. Biol. 496, 129–142 (2009).
Zhivotovsky, L. A., Underhill, P. A., Cinnioğlu, C., Kayser, M., Morar, B., Kivisild, T. et al. The effective mutation rate at Y chromosome short tandem repeats, with application to human population-divergence time. Am. J. Hum. Genet. 74, 50–61 (2004).
Wright-Robin, M. História dos Índios do Brasil (org. Carneiro da Cunha, Manuela) 253–278 (Companhia das Letras, São Paulo, SP, Brazil, 1992).
Scliar, M. O., Gouveia, M. H., Benazzo, A., Ghirotto, S., Fagundes, N. J., Leal, T. P. et al. Bayesian inferences suggest that Amazon Yunga natives diverged from Andeans less than 5000 ybp: implications for South American prehistory. BMC Evol. Biol. 14, 174 (2014).
Busby, G. B., Brisighelli, F., Sánchez-Diz, P., Ramos-Luis, E., Martinez-Cadenas, C., Thomas, M. G. et al. The peopling of Europe and the cautionary tale of Y chromosome lineage R-M269. Proc. Biol. Sci. 279, 884–892 (2012).
Bergen, A. W., Wang, C. Y., Tsai, J., Jefferson, K., Dey, C., Smith, K. D. et al. An Asian-Native American paternal lineage identified by RPS4Y resequencing and by microsatellite haplotyping. Ann Hum Genet. 63, 63–80 (1999).
Cinnioğlu, C., King, R., Kivisild, T., Kalfoğlu, E., Atasoy, S., Cavalleri, G. L. et al. Excavating Y-chromosome haplotype strata in Anatolia. Hum Genet. 114, 127–148 (2003).
Hudjashov, G., Kivisild, T., Underhill, P. A., Endicott, P., Sanchez, J. J., Lin, A. A. et al. Revealing the prehistoric settlement of Australia by Y chromosome and mtDNA analysis. Proc Natl Acad Sci U S A 104, 8726–8730 (2007).
Myres, N. M., Rootsi, S., Lin, A. A., Järve, M., King, R. J., Kutuev, I. et al. A major Y-chromosome haplogroup R1b Holocene era founder effect in Central and Western Europe. Eur J Hum Genet 19, 95–101 (2010).
Seielstad, M. T., Hebert, J. M., Lin, A. A., Underhill, P. A., Ibrahim, M., Vollrath, D. et al. Construction of human Y-chromosomal haplotypes using a new polymorphic A to G transition. Hum Mol Genet. 3, 2159–2161 (1994).
Acknowledgements
We thank Joice Pedroso, Fabiano Assunção and Sibelle T Vilaça for technical help in some experiments and analysis, and to Davidson Campos for drawing of map figures. We thank the Laboratório Multiusuário de Genômica of Universidade Federal de Minas Gerais (Brazil) for the use of the BeadXpress. We received financial support from the National Geographic Society (USA) and the National Research Council (CNPq) and Fundação de Apoio à Pesquisa de Minas Gerais (FAPEMIG) (Brazil). QA and CTS are supported by The Wellcome Trust (098051), and FRS by CNPq.
Author information
Authors and Affiliations
Consortia
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
The Genographic Consortium Li Jin, Hui Li, & Shilin Li (Fudan University, Shanghai, China); Pandikumar Swamikrishnan (IBM, Somers, New York, United States); Asif Javed, Laxmi Parida & Ajay K Royyuru (IBM, Yorktown Heights, New York, United States); R John Mitchell (La Trobe University, Melbourne, Victoria, Australia); Pierre A Zalloua (Lebanese American University, Chouran, Beirut, Lebanon); Arun Kumar, Ganesh Prasad, Ramasamy Pitchappan, Arun Varatharajan Santhakumari (Madurai Kamaraj University, Madurai, Tamil Nadu, India); R Spencer Wells and Miguel G Vilar (National Geographic Society, Washington, District of Columbia, United States); Himla Soodyall (National Health Laboratory Service, Johannesburg, South Africa); Elena Balanovska & Oleg Balanovsky (Research Centre for Medical Genetics, Russian Academy of Medical Sciences, Moscow, Russia); Jaume Bertranpetit, Marc Haber, Marta Melé, & David Comas (Universitat Pompeu Fabra, Barcelona, Spain); Christina J Adler, Alan Cooper, Clio S I Der Sarkissian & Wolfgang Haak (University of Adelaide, South Australia, Australia); Matthew E Kaplan & Nirav C Merchant (University of Arizona, Tucson, Arizona, United States); Colin Renfrew (University of Cambridge, Cambridge, United Kingdom); Andrew C Clarke & Elizabeth A Matisoo-Smith (University of Otago, Dunedin, New Zealand); Jill B Gaieski (University of Pennsylvania, Philadelphia, Pennsylvania, United States).
Supplementary Information accompanies the paper on Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Jota, M., Lacerda, D., Sandoval, J. et al. New native South American Y chromosome lineages. J Hum Genet 61, 593–603 (2016). https://doi.org/10.1038/jhg.2016.26
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2016.26
This article is cited by
-
The Pre-Columbian Peopling and Population Dispersals of South America
Journal of Archaeological Research (2021)
-
Tracing the genetic history of the ‘Cañaris’ from Ecuador and Peru using uniparental DNA markers
BMC Genomics (2020)
-
Genetic ancestry of families of putative Inka descent
Molecular Genetics and Genomics (2018)
-
The study of human Y chromosome variation through ancient DNA
Human Genetics (2017)