Abstract
Few mutations in cis have been annotated for F508del homozygous patients. Southern Italy patients who at a first analysis appeared homozygous for the F508del mutation (n=63) or compound heterozygous for the F508del and another mutation in the cystic fibrosis transmembrane conductance regulator gene (n=155) were searched for the A238V mutation in exon 6. The allelic frequency of the complex allele [A238V;F508del] was 0.04. When the whole data set was used (comprised also of 56 F508del/F508del and 34 F508del/other mutation controls), no differences reached the statistical significance in the clinical parameters, except chloride concentrations which were lower in [A238V;F508del]/other mutation compared with F508del/other mutation (P=0.03). The two study groups presented less complications than the control groups. Within the minimal data set (34 F508del/F508del, 27 F508del/other mutation, 4 [A238V;F508del]/F508del cases and 5 [A238V;F508del]/other mutation cases); that is, presenting all the variables in each patient, forced expiratory volume in 1 s and forced vital capacity presented a trend to lower levels in the study groups in comparison with the F508del/F508del group, and C-reactive protein approximated statistically significant higher levels in the [A238V;F508del]/other mutation as compared with F508del/F508del patients (P=0.09). The analysis of statistical dependence among the variables showed a significant anticorrelation between chloride and body mass index in the [A238V;F508del]/other mutation group. In conclusion, the complex allele [A238V;F508del] seems to be associated with less general complications than in the control groups, on the other hand possibly giving a worse pulmonary phenotype and higher systemic/local inflammatory response. These findings have implications for the correct recruitment and clinical response of F508del patients in the clinical trials testing the new etiological drugs for cystic fibrosis.
Similar content being viewed by others
Introduction
Cystic fibrosis (CF; MIM ID: 219700) is the most common severe autosomal-recessive genetic disorder in Caucasians and is caused by mutations in the cystic fibrosis transmembrane conductance regulator (CFTR; MIM ID: 602421; GenBank: NM_000492.3) gene. To the present day, 2006 sequence variations have been identified in the CFTR gene (http://www.genet.sickkids.on.ca/, accessed 19 November 2015). The most prevalent mutation results in deletion of phenylalanine at position 508 (c.1521_1523delCTT p.Phe508del F508del), which occurs on around 70% of CF chromosomes worldwide. With the improvement of genetic techniques, such as extensive sequencing, a large number of genetic variants has been collected, including complex alleles, with two or more mutations in cis position. As mutations in cis may act in concert to alter or reverse defective CFTR function and thus modify the CF phenotype,1 it is difficult to evaluate the contribution of each mutation or polymorphism to the phenotype. Indeed, frequently CFTR mutations or variants combined in cis may produce a more deleterious effect than each mutation alone.1, 2, 3 Therefore, it would be important to scan the entire gene in patients who are already genetically characterized, in order to find other variants which may affect the phenotype, before analyzing extra-CFTR genetic factors or considering targeted therapy to single mutations.
Despite elevated numbers of CFTR mutations detected in CF patients, in recent years the introduction of small molecule disease-modifying drugs has started the era of ‘personalized medicine’ for CF. This strategy is aimed to target the underlying defects in the CFTR protein; that is, these drugs are CFTR mutation class-specific.4 The potentiator ivacaftor, the first etiological drug for CF, has been approved for people with CF aged ⩾2 with class III CFTR mutations, while Orkambi (a combination of ivacaftor and the corrector lumacafactor) is in use in people with CF who have two copies of the F508del CFTR mutation and are aged ⩾12 years (https://tools.cff.org/research/drugdevelopmentpipeline/). The presence of genetic variants in cis may thus impact on the response to these important and novel therapeutic approaches.
We detected the new c.[713C>T;1521_1523delCTT] p.[Ala238Val;Phe508del] [A238V;F508del] complex allele in a patient with congenital bilateral absence of vas deferens (CBAVD) and in 17 patients with CF previously genetically characterized as homozygotes or compound heterozygotes for two mutations in the CFTR gene. A238V lies in the fourth transmembrane helix within the membrane spanning domain 1 of the CFTR protein, replacing an alanin by a valin at position 238. It has been first described by Picci et al. (http://www.genet.sickkids.on.ca/MutationDetailPage.external?sp=1537), who identified it in a patient aged 35 years with CBAVD in trans arrangement with F508del. The patient also had c.[1210-12[5];1210-34TG[12]] TG12-T5 (there is no protein name) variant and he was pancreatic insufficient, with moderate/mild lung disease and with sweat chloride of 60 mmol l−1. To the best of our knowledge, A238V has never been described within a complex allele.
An Italian man of 35 years with no family history of CF was referred to our laboratory after a diagnosis of CBAVD. According to a first CFTR mutational analysis, the subject appeared to be compound heterozygote for F508del and A238V mutations. He also presented the c.743+40A>G 875+40A/G (there is no protein name) variant. The genetic investigation in relatives showed the presence of both F508del and A238V in his father and in his first sister, the variant 875+40A>G in the second and no mutations in the third and fourth, so concluding that A238V was in cis with F508del. A further CFTR mutational analysis revealed the presence of c.2991G>C p.Leu997Phe L997F mutation, absent in father’s DNA. The genotype c.[713C>T;1521_1523delCTT]/[2991G>C] p.[Ala238Val;Phe508del]/[Leu997Phe] [A238V;F508del]/L997F might be consistent with the diagnosis of CFTR-related disorder (CFTR-RD) of the patient. The sweat chloride test, performed in our laboratory according to Gibson and Cooke method, was 70 mEq l−1 (normal values <60 mEq l−1), and the pancreatic exocrine function, assessed by fecal elastase measurements (>200 μg g−1), was adequate.
This case prompted us to evaluate the frequency of this new complex allele and study its effects on the phenotype of a cohort of patients from South Italy, and thereby we searched for the A238V mutation in CF patients carrying at least one F508del allele.
Materials and methods
Patients and clinical parameters
We analyzed 218 patients that at a first analysis appeared homozygous for the F508del mutation (n=63) or compound heterozygous for the F508del and another mutation in the CFTR gene (n=155). The study was approved by the ethics committee of the Azienda Ospedaliera Universitaria ‘Policlinico’ of Bari (no. 1373/CE/2012) and performed in accordance with the 1964 Declaration of Helsinki. Written informed consent from the adult study subjects or written consent from the next of kin, caretakers or guardians on behalf of the enrolled children was obtained. The chloride sweat concentration (mEq l−1) was measured at the diagnosis. Sweat test was performed using quantitative pilocarpine iontophoresis, according to classic Gibson and Cooke technique,5 in line with approved guidelines.6, 7 Reference values: normal: <30 mEq l−1, doubt borderline: 30–60 mEq l−1, pathological: >60 mEq l−1. Stimulator and clorurimetro used were, respectively: MACRODUCT SWEAT COLLECTION SYSTEM, Wescor 3700 – SYS (Wescor Inc., Logan, UT, USA) and MKII CHLORIDE ANALYZER 926S (Sherwood Scientific Ltd, Cambridge, UK). All the other clinical parameters, but not available for all patients, were the last recorded: C-reactive protein (CRP; mg dl−1), forced expiratory volume in 1 s (FEV1; % of predicted), forced vital capacity (FVC; % of predicted), and body mass index (BMI; kg m−2). Assessment of exocrine pancreatic function was performed by measuring fecal elastase-1 concentration (normal values ⩾200 μg g−1) with commercial sandwich ELISA Kit (ScheBo Biotech, Giessen, Germany) that uses two monoclonal antibodies against different specific epitopes of human pancreatic elastase.8
Sputum specimens were evaluated for microbiological tests at the first bacterial colonization. When young children could not expectorate, nasal washes and throat swab specimens were used. Processing of specimens was carried out following the Canadian CF Foundation guidelines.9 Bacterial species were identified according to the North-American guidelines.10 The following bacterial species were found: Pseudomonas aeruginosa (PA), Staphylococcus aureus (SA), Burkholderia cepacia (BC), Exophiala (Wangiella) dermatidis (ED), Serratia marcescens (SM), and methicillin-resistant Staphylococcus aureus (MRSA).
CFTR genotype analysis
At first, we looked for the A238V mutation in exon 6 in all 218 patients considered in the study with automated sequencing capillary (using BigDye Terminator version 1.1 Cycle Sequencing kit and ABI 310 automated sequencer, Applied Biosystems, Foster City, CA, USA), and then we scanned the 27 exons of the gene and their boundaries using denaturing gradient gel electrophoresis,11 followed by automated sequencing to confirm and characterize positive findings. This procedure was followed for all the patients enrolled in the study. The first time a mutation and/or a genotype and/or a complex allele appeared, it was named with three notations: nucleotidic HGVS, proteic HGVS, and legacy. Then they were called using the legacy name. The CFTR1 database was used for the three notations of each CFTR mutation or variant (http://www.genet.sickkids.on.ca/app).
In silico analysis
Prediction of functional effects of the A238V mutation was carried out using the softwares PolyPhen2 (http://genetics.bwh.harvard.edu/pph2/) and MutationTaster (http://www.mutationtaster.org/).
Statistical analysis
Our statistical analysis started from a complete data set made up of 108 patients (56 F508del/F508del controls, 34 F508del/other mutation controls, 7 [A238V;F508del]/F508del cases and 11 [A238V;F508del]/other mutation cases) measured over the 6 variables: chloride, CRP, FEV1, FVC, BMI, and pancreatic function. As not all measures were available for each variable, from these we gathered a minimal data set for the common 34 F508del/F508del and 27 F508del/other mutation controls shared by 4 [A238V;F508del]/F508del cases and 5 [A238V;F508del]/other mutation cases. Descriptive statistics were performed on each group, computing mean, median, range, s.d., s.e. and s.e./mean. This last indicator was used to show the relative variability of the data distribution within each group, when the number of replicates varies across samples.12 Only relevant values for s.e./mean in the case of CRP are shown in the paper. Because of the low number of patients in each group and as variables were not normally distributed, on this common data set we performed Mann–Whitney U-test on each group pair ([A238V;F508del]/F508del vs F508del/F508del, [A238V;F508del]/other mutation vs F508del/other mutation, [A238V;F508del]/F508del vs [A238V;F508del]/other mutation and [A238V;F508del]/other mutation vs F508del/F508del) for each variable in order to compare the median of all the possible differences between the two samples' observations. Subsequently, an analogous computation was performed on the ‘complete’ data set where, for each variable, all available values (for the reference group; that is, F508del/F508del, F508del/other mutation and study groups) were considered. We used the exact binomial test of goodness-of-fit for the variable ‘pancreatic status’ for observing whether the number of observations in each category (pancreatic sufficiency/insufficiency) fitted a theoretical expectation (controls). The choice of the exact binomial test was due to being it a nominal variable, with a small sample size. The same considerations based the use of the exact binomial test also for the comparisons about the parameter ‘bacterial colonization’. In this case, as two bacterial species (P. aeruginosa and S. aureus) were found to be common to all the four groups of patients, only these two were considered in the analysis.
For any pair of the given variables (sweat chloride, CRP, FEV1, FVC, BMI, pancreatic status), the correlation analysis tested to see whether there was an association between the two variables and how tightly the two variables were associated. Because of the not normally distribution of variables, we computed the nonparametric Spearman's rank correlation coefficient ρ (which ranges from −1 to 1) to test the statistical dependence between two variables and the related P-value (this addresses a biological question about cause-and-effect relationships between the two variables seen as reciprocally dependent). After computing all correlation coefficients among variable pairs, the strongest correlations were considered with values >0.6.13
Results
Seven of the 63 (11%) patients homozygous for F508del and 11 of the 155 (7%) compound heterozygous had A238V. The mutations present on the other allele in the compound heterozygotes with the complex allele were: c.3909C>G p.Asn1303Lys N1303K, c.1505T>C p.Ile502Thr I502T, c.4046G>A p.Gly1349Asp G1349D, c.1127_1128insA p.Gln378AlafsX4 1259insA (n=2), c.3472C>T p.Arg1158X R1158X, c.[2816A>G;2846A>T] p.[His939Arg;His949Leu] H939R;H949L, L997F, c.3883delA p.Ile1295PhefsX33 4015delA, c.1759T>A p.Phe587Ile F587I, and c.254G>A p.Gly85Glu G85E. Genetic analysis of either parents or the patients' kins confirmed that A238V was in cis configuration with the F508del mutation and consequently 18 patients in total shared the complex allele [A238V;F508del], with a allelic frequency of 0.04 (18/436). By scanning the entire gene, the following genetic variants not causing disease were found and therefore they were not considered in the statistical analysis: c.3897A>G 4029A/G, c.2562T>G 2694T/G, and c.1408A>G M470V (all these variants lack protein name).
Two programs for analyzing protein functions, Polyphen2 and MutationTaster, predicted that the A238V mutation is possibly damaging (score: 0.806, from 0.00 to 1.00) and disease causing (score: 64, from 0.0 to 215), respectively.
The 7 [A238V;F508del]/F508del patients were 3 males and 4 females (median age at diagnosis: 3.5 years); the 11 [A238V;F508del]/other mutation patients were 8 males and 3 females (median age at diagnosis: 1.8 years). As controls for the [A238V;F508del]/F508del group, we considered the F508del/F508del patients (n=56; 31 males and 25 females; median age at diagnosis: 1.2 years). As controls for the [A238V;F508del]/other mutation group, we considered the F508del/other mutation patients, which presented on the second allele the same mutations as in the [A238V;F508del]/other mutation group (n=34; 16 males and 18 females, median age at diagnosis: 2.0 years. Other mutation: N1303K=12; I502T=8; G1349D=5; R1158X=4; 4015delA=3; 1259insA=1; H939R;H949L=1). All the patients considered in this study were diagnosed as having classic CF based on an elevated sweat chloride value(>60 mEq l−1). One patient of the [A238V;F508del]/other mutation group, with the other mutation being L997F, was classified as CFTR-RD and had 70 mEq l−1 of sweat chloride.
Microbiological evaluation of sputum specimens at the moment of diagnosis identified different bacterial species in F508del/F508del patients with a prevalence for P. aeruginosa and S. aureus (Table 1). Patients with the [A238V;F508del]/F508del genotype showed equally these two bacterial species. On the other hand, F508del/other mutation patients were characterized by a net prevalence of P. aeruginosa colonization, while the [A238V;F508del]/other mutation group did not show a prevalence of one strain over the others (Table 1). As P. aeruginosa and S. aureus were common to all the groups, only these two bacterial species were included in the statistical analysis. Frequency of P. aeruginosa was neither statistically different when the two control groups were compared with their respective study group nor when the comparisons of [A238V;F508del]/other mutation vs F508del/F508del and [A238V;F508del]/F508del vs F508del/F508del were performed (Table 2). The same was found for S. aureus frequency; that is, all the comparison were not statistically significant, except for that between the [A238V;F508del]/F508del and F508del/other mutation groups (P=0.021).
Complications of the two control groups included cirrhosis, CF-related diabetes, distal intestinal obstruction syndrome, meconium ileus, pneumothorax, nontubercolosis mycobacteria and allergic bronchopulmonary aspergillosis. Interestingly, only one of the [A238V;F508del]/F508del patients presented a CF-related diabetes, and none of the [A238V;F508del]/other mutation had any complications.
The overall data set was interrogated for each considered variable (FEV1, FVC, CRP, Cl−, BMI, pancreatic status) by comparing the four different groups between them (Table 1). There were no differences that reached the statistical significance when the [A238V;F508del]/F508del group was compared with the F508del/F508del group for Cl−, CRP, FEV1, FVC and BMI. Similar results were obtained for the comparison between the [A238V;F508del]/other mutation and F508del/other mutation groups, except for Cl−, which was significantly lower in the [A238V;F508del]/other mutation patients (P=0.03). In the comparison between [A238V;F508del]/other mutation and F508del/F508del, no significant differences were found for all the considered parameters. Similarly, [A238V;F508del]/F508del and F508del/other mutation groups did not give any significant difference for all the variables. The pancreatic status was neither different between [A238V;F508del]/F508del and F508del/F508del groups nor between [A238V;F508del]/other mutation and F508del/other mutation patients (Table 1). The comparison of the pancreatic status between [A238V;F508del]/other mutation and F508del/F508del groups resulted in a non-significant difference (P=0.59), whereas the [A238V;F508del]/F508del group differed significantly from the F508del/other mutation group (P=0.04).
We then compared the clinical parameters of the study groups [A238V;F508del]/F508del and [A238V;F508del]/other mutation with those of the F508del/F508del group and F508del/other mutation groups, respectively, by using the data set presenting all the variables in each patient. This minimal data set was comprised of n=34 F508del/F508del, n=27 F508del/other mutation, n=4 [A238V;F508del]/F508del and n=5 [A238V;F508del]/other mutation (N1303K, G1349D, 1259insA, 4015delA, G85E). Bacterial colonization at the diagnosis was as follows: [A238V;F508del]/F508del: PA=2; SA=2; F508del/F508del: PA=17, SA=15, BC=1, ED=1, SM=1, MRSA=1; [A238V;F508del]/other mutation: PA=3, SA=1, BC=1; F508del/other mutation: PA=12, SA=2, BC=2. As also in this data set only PA and SA were common to all the groups, the statistical analysis of frequency of colonization was performed with these two bacterial species. Neither P. aeruginosa nor S. aureus frequency resulted in statistically significant differences among groups (Table 2).
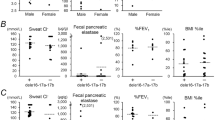
The [A238V;F508del]/F508del and the[A238V;F508del]/other mutation groups did not show any statistically significant difference in FEV1, FVC, Cl−, CRP and BMI as compared with the F508del/F508del group. However, FEV1 and FVC demonstrated a trend to decrease in the two study groups in comparison with the F508del/F508del patients (Figures 1 and 2). Moreover, in the [A238;F508del]/F508del group, CRP levels were slightly higher than those of the F508del/F508del group (Figure 1), whereas CRP was substantially higher in [A238;F508del]/other mutation in comparison with the F508del/F508 group (P=0.09), although this did not reach the statistical significance of P<0.05 (Figure 2), likely owing to the high variability in the CRP data distribution (F508del/F508del: s.e./mean=0.28; F508del/other: s.e./mean=0.18; [A238V;F508del]/F508del: s.e./mean=0.71; [A238V;F508del]/other: s.e./mean=0.43).
Cl− was slightly higher in the [A238V;F508del]/F508del group in comparison with the F508del homozygous patients (Figure 1), while it was similar in [A238V;F508del]/other mutation and F508del/F508del patients (Figure 2). The [A238V;F508del]/other mutation group did not differ in FEV1, FVC, Cl−, CRP and BMI as compared with the F508del/other mutation (Figure 3), although CRP and Cl− showed a trend to increase and decrease, respectively, with Cl− having a P-value of 0.09. FEV1, FVC, Cl−, CRP and BMI did not differ significantly between the [A238V;F508del]/F508del and the F508del/other mutation groups (Figure 4). Finally, the pancreatic status was not different between [A238V;F508del]/F508del (PI=4; PS=0) and F508del/F508del groups (PI=32; PS=2) (P=1.00), between [A238V;F508del]/other mutation (PI=4; PS=1) and F508del/other mutation groups (PI=14; PS=13) (P=0.38), between [A238V;F508del]/other mutation and F508del/F508del groups (P=0.26), and between [A238V;F508del]/F508del and F508del/other mutation groups (P=0.13).
For the minimal data set, it was possible to carry out the correlations among the six variables within each group, which showed for FEV1 and FVC a significant positive correlation in the F508del/F508del group (Table 3). Also, a significant inverse correlation between CRP and both FEV1 and FVC was found. In the study group of [A238;F508del]/F508del patients, no correlations that were strong and significant contemporarily were found (Table 3). In the [A238V;F508del]/other mutation group, the strongest and significant positive correlation was found between FEV1 and FVC; however, a strong and significant anticorrelation was also found between chloride and BMI (Table 3). A strong positive correlation between FEV1 and FVC was present in the F508del/other mutation group, as well as a strong negative and significant correlation between CRP and FEV1 and FVC, respectively (Table 3).
Discussion
The most widely used protocols for a mutational search within the CFTR gene are designed in such a way as to stop when two mutations on different alleles are found. Thus additional mutations that may be present in cis with the already found mutations may escape detection.14
This incomplete genetic characterization is common, especially for the classical forms of CF when the two mutations causing CF are identified and the analysis is stopped after they have been confirmed in parents. Certainly for this reason, few complex alleles have been described to date and not by chance especially in CFTR-RD patients, for which genetic tests are typically extended to the II and III levels. Some of these complex alleles have also been characterized by functional studies, such as c.[1727G>C;2002C>T] p.[Gly576Ala;Arg668Cys] [G576A;R668C],15, 16, 17 c.[1523T>G;3752G>A]p.[Phe508Cys;Ser1251Asn] [F508C;S1251N]18 and others.17, 19, 20, 21, 22 Mutations in cis with F508del hitherto described are c.1658G>A p.Arg553Gln R553Q and R553M (no nucleotide variant as it was being generated in vitro), and in vivo and/or in vitro tests23, 24 show that these mutations lead to a partial reversion of the phenotype. Also the alteration c.3080T>C p.Ile1027Thr I1027T of the complex allele [F508del;I1027T], very common in Britain,25 appears to retain the ability to conduct chloride, and according to CFTR2 database (http://www.cftr2.org/), it is not a CF-causing mutation.
The A238V mutation of the complex allele [A238V;F508del] identified in our laboratory has been only previously described by Picci et al. (http://www.genet.sickkids.on.ca/MutationDetailPage.external?sp=1537) in a patient with CBAVD. For the first time, here we describe it in cis arrangement with the F508del. In silico analysis performed with MutationTaster and PolyPhen2 showed that A238V has a high probability of being a mutation causing the disease, although it will be necessary to perform functional studies for a better characterization of the mutation. Indeed, none of these prediction programs reliably predicted the clinical severity of individual CFTR missense mutations with known clinical consequences.26 It is then suggested that they should not be used as diagnostic tools to predict the clinical consequences of novel missense mutations or those with unknown functional properties but just for research studies.
Based on the results obtained from the comparison of clinical data from patients F508del homozygous and compound heterozygous with the complex allele in the study, it seems that the A238V may worsen the already known phenotypic effect of the F508del mutation at the level of CRP, a systemic marker of inflammation (Figure 1). This was also found when patients with the [A238V;F508del]/other mutation genotype was compared with both having the F508del/F508del and the F508del/other mutation genotypes, respectively (Figures 2 and 3). Despite an effect of CRP increment, a statistical significance could not be evidenced owing to high variability between patients. As a speculation, thus it might be that systemic inflammation is higher in patients with the complex allele in comparison with the F508del/F508del group, possibly reflecting lung inflammation. One or more markers of pulmonary inflammation would have been indicative of this supposition.27 However, this was not confirmed by the correlation analysis shown in Table 3. Thus the two study groups show an enhancement of CRP, and so of inflammatory response, but this enhancement does not produce an appreciable effect on pulmonary function (no correlation with FEV1 and FVC) in contrary to groups without the complex allele. Nevertheless, this analysis caught an interesting statistically significant anticorrelation between chloride and BMI in the [A238V;F508del]/other mutation group. These findings are in line with a recently published report that studied whether sweat chloride concentrations can predict the clinical phenotype in CF patients.28 Among the other presented results, it is interesting to note that patients with lower sweat chloride concentrations were taller, weighed more and had a higher BMI percentile at age 12 years than patients with higher sweat chloride concentrations. It must be stressed that our observations, although in keeping with the findings just mentioned, are preliminary owing to the low number of patients available in the study groups and should be confirmed in higher numbers of patients with the [A238;F508del]/other mutation genotype.
As a consequence of this study, waiting for data on functional studies on A238V, we suggest that it would be important to change the guidelines or protocols of genetic laboratories because it is no longer sufficient to identify two mutations in trans, but it is necessary to extend genetic tests, as the presence of complex alleles could justify the phenotypic difference in patients with apparently the same genotype. In particular, in the present day, it might be important to identify any mutations in cis with the F508del mutation in the light of new drug therapies that use the combination of a potentiator (ivacaftor=VX770=Kalydeco) and a corrector (lumacaftor=VX809) directed precisely to F508del homozygous patients, whose phase II clinical trial results have been published29 and are currently in phase III clinical trials (https://tools.cff.org/research/drugdevelopmentpipeline/ and Wainwright et al.30). To the best of our knowledge, the corrector only works on the F508del mutation, and it is to be proven whether it also works well on the complex allele. The complete genotyping should be performed in order to be able to carefully select patients and prevent possible ineffectiveness of the drug or failure in the expected responses. The right selection becomes crucial for patients who are compound heterozygous for the F508del and another mutation in the CFTR gene for whom combination ivacaftor+lumacaftor therapy is still in phase II (https://clinicaltrials.gov/ct2/show/NCT01225211), so if the only copy of the F508del allele is the one present in the complex allele, the therapeutic efficacy would be questionable.
In conclusion, the complex allele [A238V;F508del] seems to determine the appearance of less general complications than in the control groups without the complex allele, on the other hand possibly giving a worse pulmonary phenotype and higher local/systemic inflammatory response.
References
Clain, J., Fritsch, J., Lehmann-Che, J., Bali, M., Arous, N., Goossens, M. et al. Two mild cystic fibrosis-associated mutations result in severe cystic fibrosis when combined in cis and reveal a residue important for cystic fibrosis transmembrane conductance regulator processing and function. J. Biol. Chem. 276, 9045–9049 (2001).
Fanen, P., Clain, J., Labarthe, R., Hulin, P., Girodon, E., Pagesy, P. et al. Structure-function analysis of a double-mutant cystic fibrosis transmembrane conductance regulator protein occurring in disorders related to cystic fibrosis. FEBS Lett. 452, 371–374 (1999).
Polizzi, A., Tesse, R., Santostasi, T., Diana, A., Manca, A., Logrillo, V. P. et al. Genotype-phenotype correlation in cystic fibrosis patients bearing [H939R;H949L] allele. Genet. Mol. Biol. 34, 416–420 (2011).
Amin, R. & Ratjen, F. Emerging drugs for cystic fibrosis. Expert Opin. Emerg. Drugs 19, 143–155 (2014).
Gibson, L. E. & Cooke, R. E. A test for concentration of electrolytes in sweat in cystic fibrosis of the pancreas utilizing pilocarpine by iontophoresis. Pediatrics 23, 545–549 (1959).
Guidelines for the Performance of the Sweat Test for the Investigation of Cystic Fibrosis in the UK (2003). http://www.acb.org.uk. Accessed 26 Jaunary 2016.
Clinical and Laboratory Standards Insitute. CLSI Document C34–A3: Sweat Testing: Sample Collection and Quantitative Analysis; Approved Guideline - Third edition. (CLSI, Wayne, PA, USA, 2009).
Stein, J., Jung, M., Sziegoleit, A., Zeuzem, S., Caspary, W. F. & Lembcke, B. Immunoreactive elastase I: clinical evaluation of a new noninvasive test of pancreatic function. Clin. Chem. 42, 222–226 (1996).
Recommendations of the Clinical Subcommittee of the Medical/Scientific Advisory Committee of the Canadian Cystic Fibrosis Foundation Microbiological processing of respiratory specimens from patients with cystic fibrosis. Can. J. Infect. Dis. 4, 166–169 (1993).
Saiman, L. & Siegel, J. Infection control recommendations for patients with cystic fibrosis: microbiology, important pathogens, and infection control practices to prevent patient-to-patient transmission. Am. J. Infect. Control 31, S1–62 (2003).
Fanen, P., Ghanem, N., Vidaud, M., Besmond, C., Martin, J., Costes, B. et al. Molecular characterization of cystic fibrosis: 16 novel mutations identified by analysis of the whole cystic fibrosis conductance transmembrane regulator (CFTR) coding regions and splice site junctions. Genomics 13, 770–776 (1992).
Eisenberg, D. T., Kuzawa, C. W. & Hayes, M. G. Improving qPCR telomere length assays: controlling for well position effects increases statistical power. Am. J. Hum. Biol. 27, 570–575 (2015).
Morabito, V. Big Data and Analytics: Strategic and Organizational Impact, (Springer International Publishing AG, Cham, Switzerland, 2015).
Lucarelli, M., Narzi, L., Pierandrei, S., Bruno, S. M., Stamato, A., d'Avanzo, M. et al. A new complex allele of the CFTR gene partially explains the variable phenotype of the L997F mutation. Genet. Med. 12, 548–555 (2010).
Pignatti, P. F., Bombieri, C., Marigo, C., Benetazzo, M. & Luisetti, M. Increased incidence of cystic fibrosis gene mutations in adults with disseminated bronchiectasis. Hum. Mol. Genet. 4, 635–639 (1995).
McGinniss, M. J., Chen, C., Redman, J. B., Buller, A., Quan, F., Peng, M. et al. Extensive sequencing of the CFTR gene: lessons learned from the first 157 patient samples. Hum. Genet. 118, 331–338 (2005).
El-Seedy, A., Girodon, E., Norez, C., Pajaud, J., Pasquet, M. C., de Becdelievre, A. et al. CFTR mutation combinations producing frequent complex alleles with different clinical and functional outcomes. Hum. Mutat. 33, 1557–1565 (2012).
Kalin, N., Dork, T. & Tummler, B. A cystic fibrosis allele encoding missense mutations in both nucleotide binding folds of the cystic fibrosis transmembrane conductance regulator. Hum. Mutat. 1, 204–210 (1992).
Wei, L., Vankeerberghen, A., Jaspers, M., Cassiman, J., Nilius, B. & Cuppens, H. Suppressive interactions between mutations located in the two nucleotide binding domains of CFTR. FEBS Lett. 473, 149–153 (2000).
Massie, R. J., Poplawski, N., Wilcken, B., Goldblatt, J., Byrnes, C. & Robertson, C. Intron-8 polythymidine sequence in Australasian individuals with CF mutations R117H and R117C. Eur. Respir. J. 17, 1195–1200 (2001).
Niel, F., Legendre, M., Bienvenu, T., Bieth, E., Lalau, G., Sermet, I. et al. A new large CFTR rearrangement illustrates the importance of searching for complex alleles. Hum. Mutat. 27, 716–717 (2006).
Masvidal, L., Igreja, S., Ramos, M. D., Alvarez, A., de Gracia, J., Ramalho, A. et al. Assessing the residual CFTR gene expression in human nasal epithelium cells bearing CFTR splicing mutations causing cystic fibrosis. Eur. J. Hum. Genet. 22, 784–791 (2014).
Dork, T., Wulbrand, U., Richter, T., Neumann, T., Wolfes, H., Wulf, B. et al. Cystic fibrosis with three mutations in the cystic fibrosis transmembrane conductance regulator gene. Hum. Genet. 87, 441–446 (1991).
Teem, J. L., Berger, H. A., Ostedgaard, L. S., Rich, D. P., Tsui, L. C. & Welsh, M. J. Identification of revertants for the cystic fibrosis delta F508 mutation using STE6-CFTR chimeras in yeast. Cell 73, 335–346 (1993).
Fichou, Y., Genin, E., Le Marechal, C., Audrezet, M. P., Scotet, V. & Ferec, C. Estimating the age of CFTR mutations predominantly found in Brittany (Western France). J. Cyst. Fibros. 7, 168–173 (2008).
Dorfman, R., Nalpathamkalam, T., Taylor, C., Gonska, T., Keenan, K., Yuan, X. W. et al. Do common in silico tools predict the clinical consequences of amino-acid substitutions in the CFTR gene? Clin. Genet. 77, 464–473 (2010).
Sagel, S. D., Wagner, B. D., Anthony, M. M., Emmett, P. & Zemanick, E. T. Sputum biomarkers of inflammation and lung function decline in children with cystic fibrosis. Am. J. Respir. Crit. Care Med. 186, 857–865 (2012).
McKone, E. F., Velentgas, P., Swenson, A. J. & Goss, C. H. Association of sweat chloride concentration at time of diagnosis and CFTR genotype with mortality and cystic fibrosis phenotype. J. Cyst. Fibros. 14, 580–586 (2015).
Boyle, M. P., Bell, S. C., Konstan, M. W., McColley, S. A., Rowe, S. M., Rietschel, E. et al. A CFTR corrector (lumacaftor) and a CFTR potentiator (ivacaftor) for treatment of patients with cystic fibrosis who have a phe508del CFTR mutation: a phase 2 randomised controlled trial. Lancet Respir. Med. 2, 527–538 (2014).
Wainwright, C. E., Elborn, J. S., Ramsey, B. W., Marigowda, G., Huang, X., Cipolli, M. et al. Lumacaftor-Ivacaftor in patients with cystic fibrosis homozygous for Phe508del CFTR. N. Engl. J. Med. 373, 220–231 (2015).
Acknowledgements
This work was funded by the Italian Ministry of Health (Law 548/93).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Diana, A., Polizzi, A., Santostasi, T. et al. The novel complex allele [A238V;F508del] of the CFTR gene: clinical phenotype and possible implications for cystic fibrosis etiological therapies. J Hum Genet 61, 473–481 (2016). https://doi.org/10.1038/jhg.2016.15
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2016.15
This article is cited by
-
High frequency of complex CFTR alleles associated with c.1521_1523delCTT (F508del) in Russian cystic fibrosis patients
BMC Genomics (2022)
-
A commentary on the novel complex allele [A238V;F508del] of the CFTR gene: clinical phenotype and possible implications for cystic fibrosis etiological therapies
Journal of Human Genetics (2016)