Abstract
Psoriasis is a complex multifactorial chronic inflammatory skin disorder involving both genetic and environmental susceptibility factors. It is strongly associated with HLA-Cw6, but several studies suggested that further genetic factors may confer additional risk. We investigated the association of two single-nucleotide polymorphisms (SNPs), rs3212227 at the 3′-untranslated region and rs7709212 located at ~6.7 kb upstream from the transcription start site of IL12B gene in a case-control study comprising 1702 individuals from India. We found both SNPs were significantly associated with psoriasis (rs7709212: odds ratio (OR)=1.37, P-value=1.09 × 10−5; rs3212227: OR=1.38, P-value=8.88 × 10−6). IL12B gene was significantly upregulated in involved skin of psoriasis patients with risk genotype carriers (rs7709212_TT and rs3212227_TT) compared with non-risk genotype carriers (rs7709212_CC and rs3212227_GG). Significantly higher serum protein concentration of IL12 was also observed among risk allele carriers compared with non-risk allele carriers irrespective of the presence of HLA-Cw6 allele. Haplotype analysis suggested significant increased risk (OR=1.50, P-value=5.01 × 10−8) to the disease when both risk alleles of IL12B were present. IL12 serum protein concentration of risk haplotype (TT-TT) carriers showed significant upregulation compared with the non-risk carriers independent of HLA-Cw6 alleles. Our data suggested the association of IL12B with the psoriasis, however no evidence was observed for the epistatic effect of IL12B with HLA-Cw6 among the psoriasis patients in India.
Similar content being viewed by others
Introduction
Psoriasis is a chronic inflammatory skin disorder that affects individuals worldwide in all populations with a variable prevalence rate of 0–11.8%.1, 2 In India, the disease incidence is around 0.44–2.8%.3, 4 The exact cause of the disease is yet to be characterized, but it is considered to be a complex multifactorial disease involving both genetic and environmental susceptibility factors.5 Previous studies suggested psoriasis to be an autoimmune disease mediated by T cells. Cross talk between the infiltrating immune cells and keratinocytes through inflammatory cytokines are suggested to cause the epidermal hyperplasia observed in psoriasis.6 Genome-wide linkage scans and association studies identified involvement of several immune-related genes, highlighted the prominent role of genetics and immune system in psoriasis pathogenesis.6, 7, 8, 9, 10, 11, 12, 13 Genetic variations in different cytokines, their receptors and antagonists are shown to have major contribution in disease predisposition. Interleukin 12 (IL12), a pro-inflammatory cytokine produced by antigen presenting cells, is increased in psoriasis.14 It regulates downstream adaptive immune response by promoting maturation of naive CD4+ T cells toward Th1-phenotype.15 IL12 is a heterodimeric cytokine, the biologically active p70 heterodimer is composed of p35 (encoded by IL12A gene) and p40 (IL12B gene) subunits.16 IL12 is mainly secreted by activated monocytes/macrophages and dendritic cells, as well as by polymorphonuclear leukocytes and keratinocytes.17 Simultaneous expression of both subunits in the same cell is required for its activity.17, 18 However, expression analysis showed that only p40 mRNA (from IL12B gene) levels were altered in psoriatic skin, whereas p35 was uniformly expressed in both normal and lesional skin of the psoriasis patients.14
A single-nucleotide polymorphism (SNP) (rs3212227) at the 3′-untranslated region (UTR) of IL12B gene was first reported from a small-scale Japanese study19 and subsequently replicated in other populations.20, 21, 22, 23 However, contradictory observation was reported among the Mestizo population of western Mexico.24 Expression pattern of IL12B was reported to alter with the risk genotype18, 25, 26, 27 or with the risk haplotype.28 In contrary, other studies have shown IL12 (p70) protein concentration was not different in serum obtained from psoriatic patients and healthy controls among the Dutch, Mestizo and South Indian populations.24, 29, 30 Another SNP (rs6887695) at the IL12B promoter was also not associated with psoriasis in Mestizo population.24 Furthermore, IL12 p70 and p40 production did not vary upon lipopolysaccharide (LPS) treatment on blood cells derived from psoriatic and healthy individuals.29 A recent study from South India among Tamil patients showed association with IL12B (rs3212227) and IL23R (rs2201841), but they did not observe any significant difference in IL12 protein concentration in the plasma of psoriasis patients having risk genotype compared with the non-risk genotypes carriers.30
HLA-Cw6 is the most strongly associated psoriasis susceptibility locus.31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42 However, only 40–80% of affected individuals/families have shown association with this allele and penetrance of this allele is only ~10%.5 This suggested that the combined effect of HLA-Cw6 with other genetic or environmental factors may be involved in disease pathogenesis. Likewise, studies have also been conducted on possible epistatic effects of IL12B and HLA-Cw6 among the psoriasis patients. Strong association of IL12B haplotype was reported, but no evidence of any statistically significant interaction between IL12B and HLA-Cw6 was observed in the Caucasian population.21 In contrary, another study on psoriasis patients from Chinese Han population showed a significantly increased (odds ratio (OR)=36) disease risk among individuals carrying both IL12B and MHC risk alleles.43 Several controversial reports of associations with IL12B genotype, epistatic effect with HLA-Cw6 as well as differential expression pattern of IL12 suggested involvement of underlying population-specific effects of IL12B gene and its role in psoriasis pathogenesis. These variable reports prompted us to conduct the present study to determine association and expression pattern of IL12B on psoriasis patients from Eastern Indian cohort.
In this study, we investigated the association of two SNPs, rs7709212 located at the ~6.7 kb upstream from the transcription start site and rs3212227 in the 3'-UTR of the IL12B gene among the psoriasis patients in Eastern part of India. We also evaluated the interaction between IL12B and HLA-Cw6 risk alleles among the psoriasis patients. Finally, we determined the functional effect of the risk alleles in terms of IL12 gene expression and serum protein concentration among the psoriasis patients with and without IL12B risk genotypes.
Materials and methods
Study population
Totally 1702 individuals with 814 psoriasis cases and 888 healthy controls from the Eastern region of India participated in the study. Mean age of psoriasis patients was 41 years (s.d.=16.00) and the mean age of disease onset was 35.07 years (s.d.=15.30), whereas control individuals had a mean age of 39.04 (s.d.=15.06). Distribution of males in cases was 67.68% whereas in controls it was 51.29%. All patients were clinically diagnosed with psoriasis and confirmed by atleast two dermatologists. Only patients categorized as having plaque or guttate psoriasis were enrolled in the study to minimize clinical heterogeneity. Patients had mild to severe psoriasis. Healthy controls were recruited after they were clinically assessed as being without psoriasis, other autoimmune disorders, systemic disorders or without a family history of psoriasis in the first and second degree relatives. Written informed consent was obtained for all patients and controls; for children younger than 18 years of age, consent was also obtained from their parents. The study was approved by the Institutional Ethics Committee for Human Research, Indian Statistical Institute, Kolkata, India and conducted according to the Declaration of Helsinki Principles. Approximately, 3 ml of blood samples were collected from each individual. About 1.5 ml was transferred to clot activator-containing tubes for serum isolation, remaining half was kept in EDTA-containing vials for DNA extraction.
HLA-Cw6 allele and IL12B SNPs typing
Peripheral blood samples were collected and genomic DNA was extracted by Qiagen DNeasy Blood/Tissue DNA isolation kit according to the manufacturer’s protocol. HLA-Cw6 typing was carried out using sequence specific PCR (SSP-PCR) as described previously42, 44 and referred to as positive when the HLA-Cw6 allele was present and negative when not present. We selected two previously reported non-coding SNPs near IL12B gene (located on 5q31-33): rs7709212,20 6696 bp upstream of IL12B gene and rs3212227 at 3′-UTR for possible association with disease. All SNPs were genotyped on a 7900HT Fast Real-Time PCR System Instrument by using allele-specific Taqman MGB probes labeled with fluorescent dyes FAM and VIC (Applied Biosystems, Foster City, CA, USA), according to the manufacturer’s protocols. Allelic discrimination was done with the ABI PRISM 7900HT SDS and the SDS 2.2.2 program (Applied Biosystems). Ten percent of the samples were run as duplicates to check for genotyping errors.
Haplotype analysis
Previous studies have shown that a larger genomic region (rs6887695–rs3212227), −60 kb 5′ to the 3′-UTR of IL12B gene was in linkage disequilibrium,20, 21 however, we limited our study to a smaller region (rs7709212–rs3212227), −6.7 kb 5′ to the 3′-UTR, which appeared to be more functionally relevant to our gene of interest. Haplotype analysis and linkage disequilibrium pattern was studied using the SHEsis online software platform (http://analysis.bio-x.cn).45
Gene expression analyses
Uninvolved and involved skin biopsies (4 mm) were obtained from 32 psoriasis patients with written consent. Eleven patients gave consent for the biopsy of both uninvolved and involved skin, and 21 patients only gave consent for biopsy of the lesional skin. Biopsy specimens were collected in RNA later (Invitrogen, Carlsbad, CA, USA) and stored at −80 °C until processing. For total RNA extraction, biopsies were snap frozen in liquid nitrogen and grinded to powder using mortar and pestle. RNA extraction was performed from 11 paired and 21 psoriatic skin tissue samples using AllPrep DNA/RNA Mini Kit (Qiagen, Hilden, Germany) following the manufacturer’s protocol. Quality of the eluted RNA was checked in Nanodrop 2000 Spectrophotometer (Thermo Scientific, Waltham, MA, USA). About 1 μg of total RNA was used for cDNA synthesis using RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific). The product was subsequently diluted and around 10 ng was finally used for each reaction. Transcripts were quantified using a 7900HT Fast Real-Time PCR system (Applied Biosystems) using Taqman probe-primers sets purchased from Applied Biosystems (IL12B Hs01011518_m1 and GAPDH Hs02758991_g1). All values were normalized to the expression of the housekeeping gene glyceraldehyde 3-phosphate dehydrogenase (GAPDH).
Enzyme-linked immunosorbent assay
Serum was collected from 120 psoriasis patients and kept in −80 °C until further processing. Enzyme-linked immunosorbent assay (ELISA) was performed for IL12 in serum obtained from 52 patients with homozygous non-risk or risk allele carrier, using IL12A Single Analyte ELISA Kit (Qiagen) following manufacturer’s protocol. Serum IL23 concentration was detected using Quantikine ELISA kit for Human IL23 (R&D Systems, Inc. Minneapolis, MN, USA) for 19 samples. The samples were replicated twice and the average of these replicates was considered for further analysis.
Statistical analysis
Case-control analysis was performed to test genetic markers for susceptibility to psoriasis. Hardy–Weinberg equilibrium was evaluated for each SNP using the χ2-test. Association for genotype and allele frequencies between cases and controls were calculated using Pearson’s χ2-test. Fisher’s exact probability test was used where expected count was less than five. Binary logistic regression analysis was carried out using R-programming. Interaction study was performed using multinomial logistic regression in SPSS software package (SPSS Inc., Chicago, IL, USA). Expression values were compared using unpaired t-test assuming unequal variance. Since the distribution of age and sex was different in cases and controls, we had also checked the association of SNPs after adjusting for age and sex using logistic regression analysis in R. Significant P-values were corrected for multiple testing using Benjamini–Hochberg multiple hypothesis testing correction in R (https://www.r-project.org/).
Results
Association of SNPs near IL12B gene
We genotyped two non-coding SNPs, rs7709212 and rs3212227 near the IL12B gene among the 814 psoriasis cases and 888 healthy individuals from the Eastern part of India. Approximately, 64% of the cases had early onset or type I (age of disease onset ⩽40 years), whereas 36% had late onset or type II (age of disease onset >40 years) form of the disease. These SNPs were individually found to be in Hardy–Weinberg equilibrium in both psoriasis cases and healthy controls (P-value >0.05). For both SNPs, the more frequent allele was observed to be the risk allele and was the same as that reported in earlier studies.20, 21, 22 Furthermore, around 89% cases and 83% controls carried atleast one copy of the risk allele.
Significant association was observed for both SNPs at the genotype level (rs7709212: P-value=9.41 × 10−5; rs3212227: P-value=5.11 × 10−5) as well as at the allele level association analysis (rs7709212: P-value=1.09 × 10−5, OR=1.37; rs3212227: P-value=8.88 × 10−6, OR=1.38) (Table 1 and Supplementary Table 1). As age and sex distribution was found to be marginally different between cases and control, we determined the association of these SNPs after adjusting for age and sex. As shown in Table 1, we found similar observations before and after adjustment. In addition, patients with early and late onset forms (type I and type II) of the disease showed similar associations for both SNPs near the IL12B gene (Supplementary Table 2), unlike those observed in the UK cohort.46 In an effort to elucidate the mode of inheritance, association pattern of both SNPs was estimated under dominant, recessive and additive models of association. Additive association model was found to be the optimal model for both SNPs (rs7709212: P-value=9.41 × 10−5; rs3212227: P-value=5.11 × 10−5), and were considered for all further genetic association analysis (Supplementary Table 3).
Linkage disequilibrium and haplotype analysis
As both SNPs had similar association patterns and are located around the same gene, we estimated the D′ and r2 values for rs7709212 located ~6.7 kb upstream and rs3212227 located at the 3′-UTR of IL12B gene. Only a moderate level of linkage (D′=0.702 and r2=0.49) was observed for these SNPs in Indian population, consistent with previous reports in North American population.20 Haplotype analysis revealed that the most frequent haplotype with the risk alleles (rs7709212_T and rs3212227_T) was significantly higher in cases (P-value=5.13 × 10−8, OR=1.50 (95% confidence interval (CI): 1.30–1.74)), whereas the non-risk combination (rs7709212_C and rs3212227_G) was significantly less found in cases (P-value=1.69 × 10−4, OR=0.74 (95% CI: 0.63–0.86)). The other two haplotypes had low frequencies and were negatively associated with disease (Table 2). The haplotype with non-risk allele for rs3212227 and risk allele for rs7709212 showed significantly low risk (OR=0.74, P-value =4.89 × 10−2).
Interaction of IL12B with HLA-Cw6 risk allele
HLA-Cw6 allele showed a strong (OR=4.56, 95% CI: 3.62–5.74) and highly significant association (P-value <2.2 × 10−16) among psoriasis patients compared with healthy controls. HLA-Cw6 allele was present in 45.2% cases, whereas only 15.3% controls had this allele (Table 3). As HLA-Cw6 had a much stronger effect on disease predisposition compared with IL12B, we wanted to determine if there was any combined effect of both these risk alleles. We considered the non-risk combination of both loci as the reference and calculated the risk imparted by different genotypes (Table 4). Presence of both HLA-Cw6 and IL12B risk genotypes significantly increased the risk of disease by 9.23 (95% CI: 5.83–14.62; P-value <2.2 × 10−16) for rs7709212 and 11.68-fold (95% CI: 7.22–18.90; P-value=<2.2 × 10−16) for rs3212227 compared with the non-risk combination. Although in absence of HLA-Cw6, IL12B risk genotypes significantly increased disease risk by 1.91-fold (95% CI: 1.31–2.78; P-value=6.83 × 10−4) for rs7709212 and 2.06-fold (95% CI: 1.40–3.05; P-value=2.32 × 10−4) for rs3212227 (Table 4). When the homozygous non-risk genotype of IL12B was considered as reference in presence of HLA-Cw6, the risk still increased ~1.5 times for the risk genotype (rs7709212: OR=1.41; rs3212227: OR=1.54) compared with the non-risk genotype, but was not significant at the level of 0.05 (Supplementary Table 4). This result indicates that irrespective of the presence or absence of HLA-Cw6, the OR increases 1.5–2 times for IL12B risk genotype compared with homozygous non-risk one (Table 4, Supplementary Table 4), and suggested lack of interaction between these two loci. Furthermore, we used multinomial logistic ratio test, including the interaction term as a covariate, which also showed no significant interaction between IL12B and HLA-C risk alleles (P-value=7.09 × 10−1 for rs7709212; P-value=9.35 × 10−1 for rs3212227) (Supplementary Table 5) among the psoriasis patients of India. When we classified the samples based on the presence of HLA-Cw6, the allele frequencies were significantly different only in HLA-Cw6-negative group. However, the ORs were similar between HLA-Cw6 present and absent samples (Supplementary Table 6). The lack of significance in Cw6-positive group can be attributed to the low number of Cw6 positive individuals among the control subjects. To determine whether IL12B and HLA-Cw6 had any joint functional effect, we further determined the expression pattern of the IL12B and its effect in the absence or presence of HLA-Cw6 risk allele.
Over expression of IL12B gene in risk allele carriers
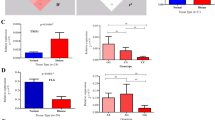
Genetic associations of IL12B SNPs are evident from our study. To establish the functional effect of the risk alleles, we studied the expression pattern of IL12B gene in the involved and uninvolved skin tissues of psoriasis patients. We performed quantitative real-time PCR from mRNAs isolated from uninvolved and involved psoriatic skin. We observed significantly high levels of IL12B mRNA expression in lesional skin compared with the uninvolved skin (fold change=19.80, P-value=3.77 × 10−7) (Figure 1a). We also observed significantly higher expression level of IL12B in patients carrying the homozygous risk genotype compared with those with homozygous non-risk genotype (rs7709212: P-value=0.02) (rs3212227: P-value=0.05) (Figures 1b, c). As our genetic association study showed that the IL12B risk haplotype was the most significantly associated haplotype with the disease, we sought to determine the expression pattern of IL12B gene with this risk haplotype, which again had a similar pattern but did not reach the level of significance at 0.05 (P-value=6.80 × 10−2) (Figure 1d).
mRNA expression pattern of IL12B. (a) IL12B mRNA expression in psoriatic and adjacent normal tissue; expression with respect to genotypes of (b) rs7709212 and (c) rs3212227, and (d) with respect to IL12B haplotypes. *P-value ⩽0.05 was considered significant. IL, interleukin. A full color version of this figure is available at the Journal of Human Genetics journal online.
Considering this aspect, we determined the protein expression in the serum of psoriasis patients harboring different genotypes. We observed significantly increased IL12 serum protein concentration for both risk genotypes (for rs7709212: P-value=4.12 × 10−5; rs3212227: P-value=1.98 × 10−4) (Figures 2a and b), as well as with the risk haplotype (P-value=1.98 × 10−4) (Figure 2c). Serum protein concentration of IL12 was significantly increased with the risk genotype compared with non-risk genotypes irrespective of the presence of HLA-Cw6 allele (Figures 2d and e). Furthermore, patients with the risk haplotype had significantly higher IL12 protein concentration in serum compared with the non-risk haplotype carriers both in the presence (P-value=3.47 × 10−4) and absence (P-value=3.24 × 10−3) of HLA-Cw6 allele (Figure 2f). Absence of epistatic interaction between HLA-Cw6 and IL12B is thus evident in terms of IL12 expression in the serum too. As p40 subunit (encoded by IL12B gene) is shared by both IL12 and IL23 cytokines, we also determined the expression of IL23 in serum of psoriasis patients. Although the expression of IL23 was higher in the risk genotype carriers compared with the non-risk genotype but was not significantly different (P-value=1.10 × 10−1) (Supplementary Figure 1).
Expression of secreted interleukin 12 (IL12) in patient serum. Protein expression of IL12 with respect to genotype of (a) rs7709212 (b) rs3212227 and (c) haplotype of IL12B; protein expression after classification on the basis of absence or presence of HLA-Cw6 allele for (d) rs7709212 (e) rs3212227 and (f) IL12B haplotype. *P-value ⩽0.05 was considered significant. A full color version of this figure is available at the Journal of Human Genetics journal online.
Discussion
In the present study, we have determined the status of genetic association of IL12B gene in the pathogenesis of psoriasis. We have analyzed the association of two SNPs, rs7709212 located upstream (−6696 bp from transcription start site) and rs3212227 in the 3′-UTR of IL12B gene among the psoriasis patients in Eastern India. IL12B was found to be strongly associated with the disease; however, this association appeared to be independent of the presence of HLA-Cw6 allele. The high similarities of genotype and allele frequencies between these SNPs suggest that they may be in strong linkage disequilibrium, but we observed relatively moderate linkage disequilibrium (r2=0.49). Further evaluation suggested that only 70% of the samples were in concordance and the remaining had discordant genotypes for these SNPs. In association analysis at both genotype and allele level, we observed significant association for both SNPs only in the psoriasis patients without HLA-Cw6 allele. However, further analysis suggested lack of any epistatic interaction between these two loci among the psoriasis patients in India. Only 15% of the normal individuals had HLA-Cw6 allele, and the low sample size in this group reduced the statistical power of the association analysis after HLA-Cw6 stratification. To elucidate the functional outcome of this genetic association, we have also observed that the expression of IL12B gene significantly correlated with the presence of risk allele in patients. We also showed that HLA-Cw6 also had no functional involvement in terms of IL12 expression. Expression quantitative trait loci (eQTL) analysis in the GTEx database (http://www.gtexportal.org/home/gene/IL12B) did not find rs7709212 or rs3212227 as an eQTL for IL12B in blood or any other tissue samples. Interestingly, association of rs3212227 with IL12B expression has also been reported in some previous studies on psoriasis,18, 26, 27 suggesting that these loci might be controlling IL12B expression only in disease conditions. This gene, therefore, appears to be an important candidate for psoriasis and these SNPs require further functional characterization. Note that none of the SNPs studied here are listed in the eQTL database of psoriatic skin, uninvolved skin and normal skin47 (http://csg.sph.umich.edu/junding/eQTL/).
Genetic association of IL12B SNPs observed in our study is in agreement with reports from south Indian population as well as with other Asian and Caucasian populations studied. This indicates that IL12B might be playing a prominent role in psoriasis pathogenesis. However, contradictory reports from The Netherlands and Mexican population point out to the inherent genetic variations and highlights the importance of population-specific studies.
Presence of epistatic interaction between HLA-Cw6 and IL12B was debatable due to conflicting reports from Caucasian21 and Chinese studies.43 Interestingly, in our study, when we considered the non-risk combination (HLA-Cw6 absent and IL12B homozygous non-risk genotypes) as reference, there was 9–11-folds increased risk in the presence of both risk alleles (HLA-Cw6 present and IL12B homozygous risk genotypes) (Table 4). However, further analysis proved this to be only the effect of HLA-Cw6. In the presence of HLA-Cw6, there was no significant increase in risk (P-value=2.98 × 10−1 for rs7709212 and P-value=2.02 × 10−1 for rs3212227) between IL12B homozygous non-risk to risk genotypes (Supplementary Table 4). When classified based on HLA-Cw6 status, both Cw6-positive and negative groups showed similar increase in risk due to IL12B risk genotype. HLA-Cw6 status also did not have any effect on serum IL12 concentrations. It is thus evident that risk imparted by IL12B to psoriasis predisposition is independent of HLA-Cw6 status in Indian population.
IL12B, encoding the p40 subunit shared between IL12 and IL23, has a prominent role in psoriasis pathogenesis is thus evident from recent studies. This fact is also supported by recent observations that a human monoclonal antibody against the p40 subunit, ustekinumab, was effective in treatment of chronic plaque psoriasis in a phase III placebo-controlled study.48 However, a recent study on Italian population did not find any association between IL12B SNP genotypes and response to ustekinumab.49 But in the presence of HLA-Cw6, there was a significant increase in the percentage of responders.49 Better response in HLA-Cw6-positive patients from ustekinumab treatment was also observed in a Chinese population,50 in agreement with the epistatic interaction observed in Chinese cohort.43 However, due to the observed absence of interaction among Indian population, association of HLA-Cw6 with ustekinumab response must be studied thoroughly before applying it in clinical practice.
In conclusion, our study showed a significant association of IL12B SNPs with psoriasis. Gene expression as well as serum protein concentration also varied with IL12B genotypes, suggesting that this gene might be playing a prominent role in psoriasis pathogenesis. However, lack of epistatic interaction with HLA-Cw6 indicates that risk imparted by IL12B to psoriasis predisposition is independent of HLA-Cw6 status in Indian population.
References
Chandran, V. & Raychaudhuri, S. P. Geoepidemiology and environmental factors of psoriasis and psoriatic arthritis. J. Autoimmun. 34, J314–J321 (2010).
Gudjonsson, J. E., Karason, A., Antonsdottir, A. A., Runarsdottir, E. H., Gulcher, J. R., Stefansson, K. et al. HLA-Cw6-positive and HLA-Cw6-negative patients with psoriasis vulgaris have distinct clinical features. J. Invest. Dermatol. 118, 362–365 (2002).
Dogra, S. & Yadav, S. Psoriasis in India: prevalence and pattern. Indian J. Dermatol. Venereol. Leprol. 76, 595–601 (2010).
Chandra, A., Ray, A., Senapati, S. & Chatterjee, R. Genetic and epigenetic basis of psoriasis pathogenesis. Mol. Immunol. 64, 313–323 (2015).
Bowcock, A. M. & Cookson, W. O. The genetics of psoriasis, psoriatic arthritis and atopic dermatitis. Hum. Mol. Genet. 13, R43–R55 (2004).
Bowcock, A. M. & Krueger, J. G. Getting under the skin: the immunogenetics of psoriasis. Nat. Rev. Immunol. 5, 699–711 (2005).
Nair, R. P., Duffin, K. C., Helms, C., Ding, J., Stuart, P. E., Goldgar, D. et al. Genome-wide scan reveals association of psoriasis with IL-23 and NF-kappaB pathways. Nat. Genet. 41, 199–204 (2009).
Zhang, X. J., Huang, W., Yang, S., Sun, L. D., Zhang, F. Y., Zhu, Q. X. et al. Psoriasis genome-wide association study identifies susceptibility variants within LCE gene cluster at 1q21. Nat. Genet. 41, 205–210 (2009).
Strange, A., Capon, F., Spencer, C. C., Knight, J., Weale, M. E., Allen, M. H. et al. A genome-wide association study identifies new psoriasis susceptibility loci and an interaction between HLA-C and ERAP1. Nat. Genet. 42, 985–990 (2010).
Stuart, P. E., Nair, R. P., Ellinghaus, E., Ding, J., Tejasvi, T., Gudjonsson, J. E. et al. Genome-wide association analysis identifies three psoriasis susceptibility loci. Nat. Genet. 42, 1000–1004 (2010).
Tsoi, L. C., Spain, S. L., Ellinghaus, E., Stuart, P. E., Capon, F., Knight, J. et al. Enhanced meta-analysis and replication studies identify five new psoriasis susceptibility loci. Nat. Commun. 6, 7001 (2015).
Yin, X., Low, H. Q., Wang, L., Li, Y., Ellinghaus, E., Han, J. et al. Genome-wide meta-analysis identifies multiple novel associations and ethnic heterogeneity of psoriasis susceptibility. Nat. Commun. 6, 6916 (2015).
Aterido, A., Julia, A., Ferrandiz, C., Puig, L., Fonseca, E., Fernandez-Lopez, E. et al. Genome-wide pathway analysis identifies genetic pathways associated with psoriasis. J. Invest. Dermatol. 136, 593–602 (2016).
Yawalkar, N., Karlen, S., Hunger, R., Brand, C. U. & Braathen, L. R. Expression of interleukin-12 is increased in psoriatic skin. J. Invest. Dermatol. 111, 1053–1057 (1998).
Trinchieri, G. Interleukin-12: a cytokine produced by antigen-presenting cells with immunoregulatory functions in the generation of T-helper cells type 1 and cytotoxic lymphocytes. Blood 84, 4008–4027 (1994).
Wolf, S. F., Temple, P. A., Kobayashi, M., Young, D., Dicig, M., Lowe, L. et al. Cloning of cDNA for natural killer cell stimulatory factor, a heterodimeric cytokine with multiple biologic effects on T and natural killer cells. J. Immunol. 146, 3074–3081 (1991).
Trinchieri, G. Interleukin-12 and the regulation of innate resistance and adaptive immunity. Nat. Rev. Immunol. 3, 133–146 (2003).
Yilmaz, V., Yentur, S. P. & Saruhan-Direskeneli, G. IL-12 and IL-10 polymorphisms and their effects on cytokine production. Cytokine 30, 188–194 (2005).
Tsunemi, Y., Saeki, H., Nakamura, K., Sekiya, T., Hirai, K., Fujita, H. et al. Interleukin-12 p40 gene (IL12B) 3'-untranslated region polymorphism is associated with susceptibility to atopic dermatitis and psoriasis vulgaris. J. Dermatol. Sci. 30, 161–166 (2002).
Cargill, M., Schrodi, S. J., Chang, M., Garcia, V. E., Brandon, R., Callis, K. P. et al. A large-scale genetic association study confirms IL12B and leads to the identification of IL23R as psoriasis-risk genes. Am. J. Hum. Genet. 80, 273–290 (2007).
Nair, R. P., Ruether, A., Stuart, P. E., Jenisch, S., Tejasvi, T., Hiremagalore, R. et al. Polymorphisms of the IL12B and IL23R genes are associated with psoriasis. J. Invest. Dermatol. 128, 1653–1661 (2008).
Capon, F., Di Meglio, P., Szaub, J., Prescott, N. J., Dunster, C., Baumber, L. et al. Sequence variants in the genes for the interleukin-23 receptor (IL23R) and its ligand (IL12B) confer protection against psoriasis. Hum. Genet. 122, 201–206 (2007).
Chang, Y. T., Chou, C. T., Yu, C. W., Lin, M. W., Shiao, Y. M., Chen, C. C. et al. Cytokine gene polymorphisms in Chinese patients with psoriasis. Br. J. Dermatol. 156, 899–905 (2007).
Sandoval-Talamantes, A. K., Brito-Luna, M. J., Fafutis-Morris, M., Villanueva-Quintero, D. G., Graciano-Machuca, O., Ramirez-Duenas, M. G. et al. The 3'UTR 1188 A/C polymorphism of IL-12p40 is not associated with susceptibility for developing plaque psoriasis in Mestizo population from western Mexico. Immunol. Lett. 163, 221–226 (2015).
Morahan, G., Huang, D., Ymer, S. I., Cancilla, M. R., Stephen, K., Dabadghao, P. et al. Linkage disequilibrium of a type 1 diabetes susceptibility locus with a regulatory IL12B allele. Nat. Genet. 27, 218–221 (2001).
Seegers, D., Zwiers, A., Strober, W., Pena, A. S. & Bouma, G. A TaqI polymorphism in the 3'UTR of the IL-12 p40 gene correlates with increased IL-12 secretion. Genes Immun. 3, 419–423 (2002).
Stanilova, S. & Miteva, L. Taq-I polymorphism in 3'UTR of the IL-12B and association with IL-12p40 production from human PBMC. Genes Immun. 6, 364–366 (2005).
Johnston, A., Xing, X., Swindell, W. R., Kochkodan, J., Riblett, M., Nair, R. P. et al. Susceptibility-associated genetic variation at IL12B enhances Th1 polarization in psoriasis. Hum. Mol. Genet. 22, 1807–1815 (2013).
Litjens, N. H., van der Plas, M. J., Ravensbergen, B., Numan-Ruberg, S. C., van Assen, Y., Thio, H. B. et al. Psoriasis is not associated with IL-12p70/IL-12p40 production and IL12B promoter polymorphism. J. Invest. Dermatol. 122, 923–926 (2004).
Indhumathi, S., Rajappa, M., Chandrashekar, L., Ananthanarayanan, P. H., Thappa, D. M. & Negi, V. S. Investigation of association of the IL-12B and IL-23 R genetic variations with psoriatic risk in a South Indian Tamil cohort. Hum. Immunol. 77, 54–62 (2016).
Enerback, C., Nilsson, S., Enlund, F., Inerot, A., Samuelsson, L., Wahlstrom, J. et al. Stronger association with HLA-Cw6 than with corneodesmosin (S-gene) polymorphisms in Swedish psoriasis patients. Arch Dermatol. Res. 292, 525–530 (2000).
Fan, X., Yang, S., Sun, L. D., Liang, Y. H., Gao, M., Zhang, K. Y. et al. Comparison of clinical features of HLA-Cw*0602-positive and -negative psoriasis patients in a Han Chinese population. Acta Derm. Venereol. 87, 335–340 (2007).
Gottlieb, A. B. & Krueger, J. G. HLA region genes and immune activation in the pathogenesis of psoriasis. Arch Dermatol. 126, 1083–1086 (1990).
Helms, C., Saccone, N. L., Cao, L., Daw, J. A., Cao, K., Hsu, T. M. et al. Localization of PSORS1 to a haplotype block harboring HLA-C and distinct from corneodesmosin and HCR. Hum. Genet. 118, 466–476 (2005).
Martinez-Borra, J., Brautbar, C., Gonzalez, S., Enk, C. D., Lopez-Vazquez, A. & Lopez-Larrea, C. The region of 150 kb telometic to HLA-C is associated with psoriasis in the Jewish population. J. Invest. Dermatol. 125, 928–932 (2005).
Pietrzyk, J. J., Turowski, G. & Kapinska-Mrowka, M. Family studies in psoriasis. II. Inheritance of HLA genotypes. Arch Dermatol. Res. 273, 295–300 (1982).
Tiilikainen, A., Lassus, A., Karvonen, J., Vartiainen, P. & Julin, M. Psoriasis and HLA-Cw6. Br. J. Dermatol. 102, 179–184 (1980).
Nair, R. P., Henseler, T., Jenisch, S., Stuart, P., Bichakjian, C. K., Lenk, W. et al. Evidence for two psoriasis susceptibility loci (HLA and 17q) and two novel candidate regions (16q and 20p) by genome-wide scan. Hum. Mol. Genet. 6, 1349–1356 (1997).
Enerback, C., Martinsson, T., Inerot, A., Wahlstrom, J., Enlund, F., Yhr, M. et al. Evidence that HLA-Cw6 determines early onset of psoriasis, obtained using sequence-specific primers (PCR-SSP). Acta Derm. Venereol. 77, 273–276 (1997).
Nair, R. P., Stuart, P. E., Nistor, I., Hiremagalore, R., Chia, N. V., Jenisch, S. et al. Sequence and haplotype analysis supports HLA-C as the psoriasis susceptibility 1 gene. Am. J. Hum. Genet. 78, 827–851 (2006).
Fan, X., Yang, S., Huang, W., Wang, Z. M., Sun, L. D., Liang, Y. H. et al. Fine mapping of the psoriasis susceptibility locus PSORS1 supports HLA-C as the susceptibility gene in the Han Chinese population. PLoS Genet. 4, e1000038 (2008).
Chandra, A., Lahiri, A., Senapati, S., Basu, B., Ghosh, S., Mukhopadhyay, I. et al. Increased risk of psoriasis due to combined effect of HLA-Cw6 and LCE3 risk alleles in Indian population. Sci. Rep. 6, 24059 (2016).
Zheng, H. F., Zuo, X. B., Lu, W. S., Li, Y., Cheng, H., Zhu, K. J. et al. Variants in MHC, LCE and IL12B have epistatic effects on psoriasis risk in Chinese population. J. Dermatol. Sci. 61, 124–128 (2011).
Bunce, M. PCR-sequence-specific primer typing of HLA class I and class II alleles. Methods Mol. Biol. 210, 143–171 (2003).
Li, Z., Zhang, Z., He, Z., Tang, W., Li, T., Zeng, Z. et al. A partition-ligation-combination-subdivision EM algorithm for haplotype inference with multiallelic markers: update of the SHEsis. Cell Res. 19, 519–523 (2009).
Smith, R. L., Warren, R. B., Eyre, S., Ho, P., Ke, X., Young, H. S. et al. Polymorphisms in the IL-12beta and IL-23R genes are associated with psoriasis of early onset in a UK cohort. J. Invest. Dermatol. 128, 1325–1327 (2008).
Ding, J., Gudjonsson, J. E., Liang, L., Stuart, P. E., Li, Y., Chen, W. et al. Gene expression in skin and lymphoblastoid cells: refined statistical method reveals extensive overlap in cis-eQTL signals. Am. J. Hum. Genet. 87, 779–789 (2010).
Leonardi, C. L., Kimball, A. B., Papp, K. A., Yeilding, N., Guzzo, C., Wang, Y. et al. Efficacy and safety of ustekinumab, a human interleukin-12/23 monoclonal antibody, in patients with psoriasis: 76-week results from a randomised, double-blind, placebo-controlled trial (PHOENIX 1). Lancet 371, 1665–1674 (2008).
Galluzzo, M., Boca, A. N., Botti, E., Potenza, C., Malara, G., Malagoli, P. et al. IL12B (p40) gene polymorphisms contribute to ustekinumab response prediction in psoriasis. Dermatology 232, 230–236 (2016).
Chiu, H. Y., Wang, T. S., Chan, C. C., Cheng, Y. P., Lin, S. J. & Tsai, T. F. Human leucocyte antigen-Cw6 as a predictor for clinical response to ustekinumab, an interleukin-12/23 blocker, in Chinese patients with psoriasis: a retrospective analysis. Br. J. Dermatol. 171, 1181–1188 (2014).
Acknowledgements
This work is supported by the Science & Engineering Research Board, DST, Govt. of India (EMR/2015/002436) and intramural research funding of Indian Statistical Institute. AC is working as a CSIR-NET SRF and is thankful to CSIR for providing the fellowship. We would like to acknowledge all volunteers who participated in the study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Chandra, A., Senapati, S., Ghosh, S. et al. Association of IL12B risk haplotype and lack of interaction with HLA-Cw6 among the psoriasis patients in India. J Hum Genet 62, 389–395 (2017). https://doi.org/10.1038/jhg.2016.139
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2016.139
This article is cited by
-
Identifying the genetic associations among the psoriasis patients in eastern India
Journal of Human Genetics (2024)
-
Epigenome-wide DNA methylation regulates cardinal pathological features of psoriasis
Clinical Epigenetics (2018)