Abstract
During the past years, several empirical and statistical models have been developed to discriminate between carriers and non-carriers of germline BRCA1/BRCA2 (breast cancer 1, early onset/breast cancer 2, early onset) mutations in families with hereditary breast or ovarian cancer. Among these, the BRCAPRO or CaGene model is commonly used during genetic counseling, and plays a central role in the identification of potential carriers of BRCA1/2 mutations. We compared performance and clinical applicability of BRCAPRO version 5.1 vs version 6.0 in order to assess diagnostic accuracy of updated version. The study was carried out on 517 pedigrees of patients with familial history of breast or ovarian cancer, 150 of which were submitted to BRCA1/2 mutation screening, according to BRCAPRO evaluation or to criteria based on familial history. In our study, CaGene 5.1 was more sensitive than CaGene 6.0, although the latter showed a higher specificity. Both BRCAPRO versions better discriminate BRCA1 than BRCA2 mutations. This study evidenced similar performances in the two BRCAPRO versions even if the CaGene 6.0 has underestimated the genetic risk prediction in some BRCA mutation-positive families. Genetic counselors should recognize this limitation and during genetic counseling would be advisable to use a set of criteria in order to improve mutation carrier prediction.
Similar content being viewed by others
Introduction
Breast cancer (BC) and ovarian cancer (OC) are considered among the most common and important diseases affecting women worldwide, at present accounting for one-third of all female cancers.1 Population statistics indicated 25% of breast cancer cases are diagnosed before the age of 50 years, and 10% of these cases is carrier of a BRCA1 (breast cancer 1, early onset) or BRCA2 (breast cancer 2, early onset) mutation.2, 3, 4, 5, 6 Deleterious mutations affecting these genes cause a considerably increased risk of BC and OC as compared with the general population.7, 8 In fact, carriers of BRCA1 mutations up to 70 years old have 56–80% lifetime risk of developing BC, whereas OC risk differs by gene, being higher among BRCA1 than among BRCA2 carriers (28–66% vs 16–27%).9, 10, 11, 12 In affected patients, the identification of a BRCA1/2 mutation indicates an increased risk for a second cancer and represents an important step in the prevention of BC/OC, considerably reducing their morbidity and mortality. Unaffected relatives can undergo testing for the known mutation and clarify their own personal risks. Women who result positive for BRCA1/2 mutations can choose different prophylactic strategies, including mastectomy and/or oophorectomy, to dramatically reduce the risk of developing these cancers.13, 14, 15, 16 Unfortunately, genetic testing is expensive and time consuming, making it important to accurately select by genetic counseling patients at high risk of being a BRCA1/2 mutation carrier (carrier probability (CP)) to submit to molecular analysis. The American Society of Clinical Oncology has suggested that genetic testing should be restricted only to individuals who present a CP of >10%.17, 18 In this view, over the past years, many statistical models (such as BOADICEA, BRCAPRO (also called CaGene), Myriad and Couch (also known as PENN) have been designed to calculate the CP and to select the individuals to submit to genetic testing.19, 20, 21, 22 BRCAPRO model is a commonly used computer program able to estimate the individual CP based on family history, age at disease onset and presence of multiple tumors.23, 24, 25, 26, 27, 28, 29, 30 Several studies have evaluated the accuracy of BRCA1/2 mutation prediction models in a large cohort of patients with different ethnic backgrounds and genders.30, 31, 32, 33 However, when used by genetic counselors during the editing of the familiar pedigree, these models show some limitation, such as the impossibility to include data about ethnicity, tumor markers, multiple tumors and, most of all, information about probands’ second- and third-degree relatives. For this reason, during the time, the BRCAPRO program has undergone several improvements and changes related to possibility to add major information in the pedigree analysis. Recently, BRCAPRO version 5.1 has been updated to version 6.0 that allows the insertion in the pedigree of the maternal and paternal cousins and enables to specify in detail other risk factors such as height, weight and mammographic density. The purpose of the present study is twofold: (1) to compare performance and sensitivity of BRCAPRO 5.1 to version 6.0 and (2) to determine whether the sensitivity and specificity of the new version could suggest its usefulness during routinely genetic counseling of families with recurrent BC/OC.
Materials and methods
Participants
We retrospectively collected data from pedigrees of families recruited between 2000 and 2013 in two centers in central Italy, namely the Human Genetics division of the Pescara Hospital and the Medical Genetics service of the Chieti University. All patients provided written informed consent. All patients gave written informed consent to publication of the identifiable data within Tables 2 and 3. Ethics committee approval was not needed because all investigations performed were part of standardized routine diagnostic. In total, 517 probands, represented by patients affected by BC, OC or other cancers, or healthy subjects with BC/OC family history, underwent genetic counseling in which personal and familiar details were collected including current age, age at diagnosis, type of cancer and presence of other affected members in family. In order to avoid the bias induced by the presence of more affected members in the earlier collected families as compared with the more recent ones because of the appearance of novel cases in the former families, familiar pedigrees were not updated during the course of the study and only the original pedigrees were considered.
Risk assessment for genetic mutations
Genetic risk prediction of the 517 pedigrees was calculated with BRCAPRO model by using CaGene version 5.1 and 6.0 software packages. The probands were classified as ‘BRCAPRO positive’ (CP ⩾10%) or ‘BRCAPRO negative’ (CP <10%). All ‘BRCAPRO-positive’ probands were enrolled for molecular analysis. To investigate the presence of false ‘BRCAPRO negatives’, we classified the families for referral to BRCA genetic testing, considering the following criteria (high CP risk vs low CP risk): (1) BC before 40 years; (2) OC before 47 years; (3) bilateral BC before 43 years; (4) BC and OC before 52 years; (5) bilateral BC and OC before 56 years; and (6) male BC at any age.25
BRCA1 and BRCA2 mutation analysis
BRCA1/2 genetic testing was performed on selected probands after obtaining written informed consent to undergo genetic testing and to receive their results. For each selected family a proband (as a rule an affected patient) was chosen for molecular analysis. When no affected member was available, a first-degree unaffected relative was analyzed. Genomic DNA was extracted automatically from peripheral blood lymphocytes using BioRobot EZ1instrument (Qiagen, Milan, Italy) according to the manufacturer’s protocol. Amplification of all coding exons and of each flanking intron of BRCA1/2 genes was performed in B-Pure EasySeq PCR plates (Nimagen BV, Nijmegen, The Netherlands) followed by direct DNA sequencing. Sequence variants were recorded as BRCA1/2 mutations or unclassified/unknown variants according to the Breast Cancer Information core (BIC) database. Carriers of nondeleterious mutations or variants of uncertain clinical significance were considered as not mutated. In BRCAPRO-positive patients not carriers of BRCA1/2 sequence mutations, the presence of large genomic rearrangements was tested using the SALSA P002B-BRCA1 and P045-BRCA2 MLPA kits (MRC-Holland, Amsterdam, The Netherlands) as previously described.27
Statistical analysis
Statistical analysis was performed using SPSS package computer program version 22.0 and MedCalc software package version 13. The sensitivity, specificity, positive predictive value (PPV), negative predictive value, positive likelihood ratio and negative likelihood ratio were calculated for the CaGene 5.1 and 6.0 versions at the 10% CP threshold and with 95% confidence interval (CI). Receiver operating characteristic curves were constructed by plotting the sensitivity (true positives) against 1 minus specificity (true negatives) for all possible values of the mutation probability. The areas under the curves (AUCs) were calculated for the entire group of probands with either pathogenic mutation or a wild-type genotype. AUC associated with 95% CI is a combined measure of sensitivity and specificity and allows to discriminate between mutation carriers and those without a BRCA mutation. This area directly represents the overall accuracy of the program in identification of germline mutation carriers. As the AUC is a portion of the area of the unit square, its value will always be between 0 and 1. An AUC of 0.5 indicates random performance, and 1 denotes perfect performance. Specifically, values 0.9–1 indicate excellent predictive accuracy, values 0.8–0.9 good accuracy, values 0.7–0.8 fair accuracy, values 0.6–0.7 poor accuracy and 0.5–0.6 unacceptable accuracy.34, 35
Results
Patient selection
Out of the 517 subjects submitted to genetic counseling, 150 (29%) were selected for molecular analysis during our study period. Briefly, patients were selected if: (1) ‘BRCAPRO’ positive after risk evaluation with CaGene 5.1 or 6.0 (CP ⩾10%) (55 patients), or (2) entering in the high CP risk category based on pedigree analysis, although being BRCAPRO negative for both CaGene 5.1 and 6.0 (95 patients). All patients were female except 10 (5 males with prostate cancer, 3 with BC and 2 healthy relatives of BC patients). The 150 probands were represented by: 112 BC (of which 16 bilateral BC, 2 BC+OC and 4 with recurrent cancer in the same breast) (74.6%); 13 OC (of which 2 BC+OC and 1 bilateral BC+OC) (8.6%); 5 males with prostate cancer and BC family history (3.33%); 1 female patient with thyroid cancer and BC family history (0.66%) and, finally, 19 healthy subjects (17 females and 2 males) belonging to families with high incidence of BC and/or OC (at least one first-degree affected relative in 15 cases and at least 3 cases in 4 cases), in which no affected patient was available for analysis (12.6%) (Table 1). The average age at diagnosis in the group of selected probands affected by BC or OC was 45.21 years (range 22–77) and 42.69 years (range 19–60), respectively. The age at diagnosis of BC was <40 years in 47 (41.96%) patients, whereas the age at diagnosis of OC was <40 years in 2 (15.38%) patients.
BRCA1 and BRCA2 mutational analysis
Overall, 21 (14%) out of 150 tested individuals had BRCA mutation, of which 16 (76.2%) were in BRCA1 and 5 (238%) in BRCA2. Among patients with BRCA1 mutations, two were compound heterozygotes for BRCA1 variants considered as pathogenic according to previous literature data25, 36 (Table 2). The majority of the detected mutations (13/21, 61.9%) produced a truncated protein and are considered pathogenic in the BIC database. The most common mutations were E1373X and C61G of BRCA1, observed in four and two unrelated families, respectively. Out of the 16 BRCA1 mutations, 5 (31.25%) were found in patients with BC, 3 (18.75%) in patients with bilateral BC, 2 (12.5%) in patients with BC+OC, 3 (18.75%) in patients with OC and 3 (18.75%) in healthy subjects. On the other hand, the 5 BRCA2 mutations were observed in 2 probands with bilateral BC (40%), 2 (40%) patients with BC and 1 (20%) male patient with prostate cancer (Table 2). Variants of uncertain significance were identified in 55 cases.
Comparative performance of CaGene version 5.1 with 6.0 software packages
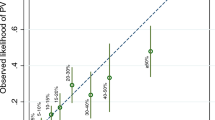
A total of 150 probands have been analyzed with both BRCAPRO versions. Fifty-two (34.66%) cases were positive for CaGene 5.1 (17 for BRCA1, 5 for BRCA2 and 30 for both) and 47 (31.33%) for CaGene 6.0 (23 for BRCA1, 5 for BRCA2 and 19 for both) (Table 3). The overall number of patients submitted to molecular analysis being positive for CaGene 5.1, CaGene 6.0 or both was 55. BRCA1 mutations were found in 16/47 (34.04%) CaGene 5.1-positive cases and in 15/42 (35.71%) CaGene 6.0-positive cases. As to BRCA2, 3/35 CaGene-5.1 positive (8.57%) and 2/24 CaGene 6.0-positive subjects (8.33%) were mutation carriers (Table 3). Of the 44 probands positive for both CaGene versions, 21 showed a mutation (47.72%), involving in 16 cases BRCA1 (36.36%) and in 5 cases BRCA2 (11.36%). Mutations were identified in 19/52 CaGene 5.1-positive cases (36.53% ) and 17/47 CaGene 6.0-positive cases (36.17%). The overall mutation detection rate in patients who were BRCAPRO positive for CaGene 5.1, 6.0 or both was 19/55 (34.54%). In only 4 cases of ‘BRCAPRO-negative’ patients (all negative for CaGene 6.0 and 2 also for CaGene 5.1), genetic testing detected a mutation, affecting BRCA2 in 3 cases and BRCA1 in the last one (Figure 1). In addition, we reported in Table 4 the real CP value calculated by CaGene 5.1 and 6.0 in patient carriers of BRCA1/2 mutations (Table 4a). The results of this analysis suggested different cutoff points for carriers of BRCA1/2 mutations (Table 4b). The calibration of the two BRCAPRO versions was assessed by statistical analysis using SPSS (version 22) and MedCalc statistical software (version 13) at 10% CP threshold. The two versions showed similar PPV for BRCA1 mutations (PPV=38% for CaGene 5.1 and PPV=36% for CaGene 6.0) (Table 5). Furthermore, both CaGene 5.1 and CaGene 6.0 presented the same PPV (9%) for BRCA2 mutations. The negative predictive value for BRCA1 mutations was also similar (1 for CaGene 5.1 and 0.99 for CaGene 6.0, respectively), and negative predictive value for BRCA2 mutations was equal for both versions (0.98) (Table 5). The positive likelihood ratio for BRCA1 mutation carriers was 4.62 (95% CI 3.35–6.38) for CaGene 5.1 and 5.23 (95% CI 3.65–7.28) for CaGene 6.0, whereas the negative likelihood ratios were consistently <0.1 (CaGene 5.1= 0.0; CaGene 6.0=0.08), indicating that both versions are capable to include mutation carrier status and to exclude false negatives. Smaller values were obtained from the positive likelihood ratio for BRCA2 mutation carriers: 2.72 (95% CI 3.35–6.38) for CaGene 5.1 and 2.90 (95% CI 3.65–7.28). Moreover, negative likelihood ratios for BRCA2 mutation carriers were considerably >0.1 (CaGene 5.1=0.51; CaGene 6.0=0.7), indicating that both versions could exclude mutation carrier status in the presence of a mutation and thus include false negatives. The CaGene 5.1 showed a higher sensitivity (95% CI) than 6.0 version for the detection of BRCA1 mutation carriers (100% vs 93.7%), whereas the specificity increased in the updated version (78% vs 82.1%). The same situation occurred with regard to BRCA2 mutation, as the sensitivity was greater in the 5.1 version (60% vs 40%), whereas the specificity was considerably higher (77.9% vs 84.8%) in the 6.0 version. These data were also confirmed by combining BRCA1/2 mutations (Table 5). Finally, the accuracy of the two BRCAPRO versions was also compared by calculating the areas under the receiver operating characteristic curves. The AUC results, listed in the Table 6, revealed that both 5.1 and 6.0 versions are able to select BRCA1 better than BRCA2 mutation carriers (Table 6 and Figure 2).
BRCA mutation-positive family. At the top are shown the probabilities generated by CaGene 5.1 and 6.0. Under each individual are described year of birth and age of diagnosis of cancer. The proband is the person tested for BRCA mutation. (a, b) The probands 46 and 63 were negative for both CaGene 5.1 and CaGene 6.0 for BRCA2 but were carriers of a mutation in this gene. (c) The proband 141 was positive for CaGene 5.1 and negative for CaGene 6.0 for BRCA2 gene but was carrier of a BRCA2 mutation. (d) The proband 125 was positive for CaGene 5.1 and negative for CaGene 6.0 for the BRCA1 gene but was carrier of a BRCA1 mutation.
Discussion
Genetic counseling and testing are increasingly integrated in the clinical management of individuals with a relevant family history of BC and/or OC.37, 38, 39 During the past several years, different probability models have been used for identifying high-risk cases to submit to molecular testing for the identification of BRCA1/2 mutations. The performance evaluation of these prediction models has a relevant impact in the clinical practice, allowing to dramatically reduce the number of genetic tests carried out on families without BRCA mutations, and increasing the cost/benefit ratio of the analysis. In this view, another crucial point is represented by adverse psychological reactions generated by the waiting for the response of genetic testing.40 In fact, the BRCA1/2 molecular test involves not only the analyzed person, but also other family members who may be at risk, thus increasing the anxiety levels in both the tested proband and his/her relatives. For this reason, an accurate selection of probands to submit to genetic testing would avoid anxiety, depression and nervousness related to the test in low-risk women.41, 42 On the other hand, the use of too stringent selection criteria would increase the number of mutation carriers not submitted to the test, preventing awareness of mutation status and subsequent access to appropriate clinical options. In the present study, we carried out a retrospective analysis on a sample of 517 subjects to evaluate the performance and clinical efficiency of two BRCAPRO versions in risk prediction for BRCA1/2 mutations, namely CaGene 5.1 and CaGene 6.0. This latter version of the software allows to overcome some practical limitations of the previous versions by allowing to incorporate into the calculations for predicting CP the cousins of the affected proband and other personal risk factors, such as reproductive history. Of the 517 analyzed subjects, 150 underwent BRCA1/2 molecular analysis, and an overall mutation detection rate of 14% (21 out of 150 analyzed patients) was detected that is lower than the one reported by the Italian Consortium of Hereditary Breast and Ovarian Cancer (23%).43 This discrepancy is likely related to the presence within our sample of 95 probands who were ‘BRCAPRO negative’ when using CaGene 5.1 and 6.0, but who were included in the study in order to provide a portion of the sample selected with low stringent criteria for the evaluation of possible false negatives generated by BRCAPRO analysis. In fact, when considering only cases selected using CaGene 5.1 or 6.0, the detection rate of our analysis increased up to 34.54% (19/55), according to our previous reports,25, 27 and confirming that the detection rate in cases selected with BRCAPRO program is ∼1 out of 3 analyzed patients, with a very positive cost/benefit ratio. In this view, CaGene 5.1 and CaGene 6.0 showed similar performances, although the new version appears to underestimate the genetic risk in some BRCA mutation-positive families. In fact, in a few cases, although we are far from defining a statistically significant difference, a discrepancy between the probabilities generated by the two versions of the BRCAPRO program and the results of genetic testing was observed. CaGene 6.0 produced a CP of <10% in four families in which molecular analysis subsequently evidenced the presence of BCRA1/2 mutations (described in Figure 1). These results are consistent with previously published studies evidencing that BRCAPRO model can fail to correctly identify the CP in families with affected males or with patients showing multiple tumors or relapses.32, 44 It is important to underline that our study has a number of limitations that should be taken into account, including its retrospective nature and small sample size. Moreover, our data also suggest that CaGene 5.1 and in particular CaGene 6.0 can underestimate the CP probability in families with affected members aged >40 years. In our sample this also occurred in families with bilateral BC, differently from previous reports.45, 46 Regarding the accuracy of the CaGene 5.1 and 6.0, we observed an equal ability to discriminate between carriers and noncarriers of BRCA1/2 mutations and to identify the most likely mutated gene. In fact, the receiver operating characteristic curves of the two models (0.852 for CaGene 5.1 and 0.879 for CaGene 6.0) were higher than the curves observed by Euhus et al.18 (0.712), Antoniou et al.7 (0.76) and Marroni et al.44 (0.757) using previous versions of the program. Moreover, although this is not strictly mean statistically significant difference, the two versions were enough comparable in terms of sensitivity and specificity. In fact, it is possible to note only slight, not significant, differences among the most clinically useful parameters, such as sensitivity and specificity: CaGene 5.1 showed a slightly higher sensitivity than 6.0 version for the detection of BRCA1/2 mutation carriers (90.5% vs 85.7%), whereas the specificity increased in the updated version (71% vs 67%). From a clinical point of view, sensitivity can be considered as more important than specificity, because it would reduce the number of false negatives. Meanwhile, high specificity would play an important role in the reduction of costs for molecular analysis and in the preservation of limited health-care resources. In fact, using CaGene 5.1 alone, we would have avoided ∼65% of the tests, but we would have missed ∼1 every 2.5 carriers of BRCA2 mutation. On the other hand, using CaGene 6.0 alone, we would have avoided 68% of the tests, but 1 every 16 BRCA1 mutation carriers and 1 every 1.6 BRCA2 mutation carriers would have been missed. In conclusion, our study suggests that the CaGene 6.0 version of the BRCAPRO model has similar performance to the previous CaGene 5.1 version, but its use in routine clinical practice and its initial validation should be performed with caution, as this program still has several limitations such as underestimation of CP in pedigrees presenting with male patients affected by cancers different from BC, female patients with bilateral breast cancer and/or multiple tumors. Genetic counselors should recognize these limitations and during genetic counseling it would be advisable to construct in detail the pedigrees of probands, to scrupulously collect the clinical histories of patients and to use a set of criteria in order to improve mutation carrier prediction. When supported by an accurate evaluation of clinical data, familial history and biological markers, CaGene 6.0 could likely represent a valuable tool to select cases of BC and/or OC for mutation analysis, allowing a more targeted management of families considered as high risk, with important implications on health-care costs.
References
Karami, F. & Mehdipour, P. A comprehensive focus on global spectrum of BRCA1 and BRCA2 mutations in breast cancer. Biomed Res. Int. 2013, 928562 (2013).
Kwon, J. S., Gutierrez-Barrera, A. M., Young, D., Sun, C. C., Daniels, M. S., Lu, K. H. et al. Expanding the criteria for BRCA mutation testing in breast cancer survivors. J. Clin. Oncol. 28, 4214–4220 (2012).
Frank, T. S., Deffenbaugh, A. M., Reid, J. E., Hulick, M., Ward, B. E., Lingenfelter, B. et al. Clinical characteristics of individuals with germline mutations in BRCA1 and BRCA2: analysis of 10,000 individuals. J. Clin. Oncol. 20, 1480–1490 (2002).
Antoniou, A., Pharoah, P. D., Narod, S., Risch, H. A., Eyfjord, J. E., Hopper, J. L. et al. Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case series unselected for family history: a combined analysis of 22 studies. Am. J. Hum. Genet. 72, 1117–1130 (2003).
Nelson, H. D., Huffman, L. H., Fu, R. & Harris, E. L. Genetic risk assessment and BRCA mutation testing for breast and ovarian cancer susceptibility: systematic evidence review for the U.S. Preventive Services Task Force. Ann. Intern. Med. 143, 362–379 (2005).
Christinat, A. & Pagani, O. Practical aspects of genetic counseling in breast cancer: lights and shadows. Breast 22, 375–382 (2013).
Antoniou, A. C., Pharoah, P. D., McMullan, G., Day, N. E., Stratton, M. R., Peto, J. et al. A comprehensive model for familial breast cancer incorporating BRCA1, BRCA2 and other genes. Br. J. Cancer. 86, 76–83 (2002).
DeJong, M. M., Nolte, I. M., te Meerman, G. J., van der Graaf, W. T. A., Oosterwijk, J. C., Kleibeuker, J. H. et al. Genes other than BRCA1 and BRCA2 involved in breast cancer susceptibility. J. Med. Genet. 39, 225–242 (2002).
Brose, M. S., Rebbeck, T. R., Calzone, K. A., Stopfer, J. E., Nathanson, K. L. & Weber, B. L. Cancer risk estimates for BRCA1 mutation carriers identified in a risk evaluation program. J. Natl. Cancer Inst. 94, 1365–1372 (2002).
Easton, D. F., Hopper, J. L., Thomas, D. C., Antoniou, A., Pharoah, P. D., Whittemore, A. S. et al. Breast cancer risks for BRCA1/2 carriers. Science 306, 2187–2191 (2004).
Wacholder, S., Struewing, J. P., Hartge, P., Greene, M. H. & Tucker, M. A. Breast cancer risks for BRCA1/2 carriers. Science 306, 2187–2191 (2004).
Laraqui, A., Uhrhammer, N., Lahlou-Amine, I., El Rhaffouli, H., El Baghdadi, J., Dehayni, M. et al. Mutation screening of the BRCA1 gene in early onset and familial breast/ovarian cancer in Moroccan population. Int. J. Med. Sci. 10, 60–67 (2013).
Rebbeck, T. R., Friebel, T., Lynch, H. T., Neuhausen, S. L., van 't Veer, L., Garber, J. E. et al. Bilateral prophylactic mastectomy reduces breast cancer risk in BRCA1 and BRCA2 mutation carriers: the PROSE Study Group. J. Clin. Oncol. 22, 1055–1062 (2004).
Kang, H. H., Williams, R., Leary, J., k ConFab Investigators, Ringland, C., Kirk, J., Ward, R. et al. Evaluation of models to predict BRCA germline mutations. Br. J. Cancer. 95, 914–920 (2006).
Tuttle, T. M., Abbott, A., Arrington, A. & Rueth, N. The increasing use of prophylactic mastectomy in the prevention of breast cancer. Curr. Oncol. Rep. 12, 16–21 (2010).
Pocobelli, G., Chubak, J., Hanson, N., Drescher, C., Resta, R., Urban, N. et al. Prophylactic oophorectomy rates in relation to a guideline update on referral to genetic counseling. Gynecol. Oncol. 126, 229–235 (2012).
American Society of Clinical Oncology. American Society of Clinical Oncology policy statement update: genetic testing for cancer susceptibility. J. Clin. Oncol. 21, 2397–2406 (2003).
Euhus, D. M., Smith, K. C., Robinson, L., Stucky, A., Olopade, O. I., Cummings, S. et al. Pretest prediction of BRCA1 or BRCA2 mutation by risk counselors and the computer model BRCAPRO. J. Natl. Cancer Inst. 94, 844–851 (2002).
James, P. A., Doherty, R., Harris, M., Mukesh, B. N., Milner, A., Young, M. A. et al. Optimal selection of individuals for BRCA mutation testing: a comparison of available methods. J. Clin. Oncol. 24, 707–715 (2006).
Oros, K. K., Ghadirian, P., Maugard, C. M., Perret, C., Paredes, Y., Mes-Masson, A. M. et al. Application of BRCA1 and BRCA2 mutation carrier prediction models in breast and/or ovarian cancer families of French Canadian descent. Clin. Genet. 70, 320–329 (2006).
Parmigiani, G., Chen, S., Iversen, E. S. Jr, Friebel, T. M., Finkelstein, D. M., Anton-Culver, H. et al. Validity of models for predicting BRCA1 and BRCA2 mutations. Ann. Intern. Med. 147, 441–450 (2007).
Jacobi, C. E., de Bock, G. H., Siegerink, B. & van Asperen, C. J. Differences and similarities in breast cancer risk assessment models in clinical practice: which model to choose? Breast Cancer Res. Treat. 115, 381–390 (2009).
Berry, D. A., Iversen, E. S. Jr, Gudbjartsson, D. F., Hiller, E. H., Garber, J. E., Peshkin, B. N. et al. BRCAPRO validation, sensitivity of genetic testing of BRCA1/BRCA2, and prevalence of other breast cancer susceptibility genes. J. Clin. Oncol. 20, 2701–2712 (2002).
Domchek, S. M., Eisen, A., Calzone, K., Stopfer, J., Blackwood, A. & Weber, B. Application of breast cancer risk prediction models in clinical practice. J. Clin. Oncol. 21, 593–601 (2003).
Stuppia, L., Di Fulvio, P., Aceto, G., Pintor, S., Veschi, S., Gatta, V. et al. BRCA1 and BRCA2 mutations in breast/ovarian cancer patients from central Italy. Hum. Mutat. 22, 178–179 (2003).
Evans, D. G., Eccles, D. M., Rahman, N., Young, K., Bulman, M., Amir, E. et al. A new scoring system for the chances of identifying a BRCA1/2 mutation outperforms existing models including BRCAPRO. J. Med. Genet. 41, 474–480 (2004).
Veschi, S., Aceto, G., Scioletti, A. P., Gatta, V., Palka, G., Cama, A. et al. High prevalence of BRCA1 deletions in BRCAPRO-positive patients with high carrier probability. Ann. Oncol. 18, 86–92 (2007).
Biswas, S., Tankhiwale, N., Blackford, A., Barrera, A. M., Ready, K., Lu, K. et al. Assessing the added value of breast tumor markers in genetic risk prediction model BRCAPRO. Breast Cancer Res. Treat. 133, 347–355 (2012).
Biswas, S., Atienza, P., Chipman, J., Hughes, K., Barrera, A. M., Amos, C. I. et al. Simplifying clinical use of the genetic risk prediction model BRCAPRO. Breast Cancer Res. Treat. 139, 571–579 (2013).
Varesco, L., Viassolo, V., Viel, A., Gismondi, V., Radice, P., Montagna, M. et al. Performance of BOADICEA and BRCAPRO genetic models and of empirical criteria based on cancer family history for predicting BRCA mutation carrier probabilities: a retrospective study in a sample of Italian cancer genetics clinics. Breast 22, 1130–1135 (2013).
Kwong, A., Wong, C. H., Suen, D. T., Co, M., Kurian, A. W., West, D. W. et al. Accuracy of BRCA1/2 mutation prediction models for different ethnicities and genders: experience in a southern Chinese cohort. World J. Surg. 36, 702–713 (2012).
Van Harssel, J. J., van Roozendaal, C. E., Detisch, Y., Brandão, R. D., Paulussen, A. D., Zeegers, M. et al. Efficiency of BRCAPRO and Myriad II mutation probability thresholds versus cancer history criteria alone for BRCA1/2 mutation detection. Fam. Cancer 9, 193–201 (2010).
Schneegans, S. M., Rosenberger, A., Engel, U., Sander, M., Emons, G. & Shoukier, M. Validation of three BRCA1/2 mutation-carrier probability models Myriad, BRCAPRO and BOADICEA in a population-based series of 183 German families. Fam. Cancer 11, 181–188 (2012).
Metz, C. E. Basic principles of ROC analysis. Semin. Nucl. Med. 8, 283–298 (1978).
Somoza, E., Soutullo-Esperon, L. & Mossman, D. Evaluation and optimization of diagnostic tests using receiver operating characteristic analysis and information theory. Int. J. Biomed. Comput. 24, 153–189 (1989).
Hadjisavvas, A., Adamou, A., O'Dowd Phanis, C., Todd, C. M., Kitsios, P., Kyriacou, K. et al. Q356R and S1512I are BRCA1 variants that may be associated with breast cancer in a Cypriot family. Oncol. Rep. 9, 383–386 (2002).
Robson, M. & Offit, K. Clinical practice. Management of an inherited predisposition to breast cancer. N. Engl. J. Med. 357, 154–162 (2007).
Meiser, B., Tucker, K., Friedlander, M., Barlow-Stewart, K., Lobb, E., Saunders, C. et al. Genetic counselling and testing for inherited gene mutations in newly diagnosed patients with breast cancer: a review of the existing literature and a proposed research agenda. Breast Cancer Res. 10, 216 (2008).
Pal, T. & Vadaparampil, S. T. Genetic risk assessments in individuals at high risk for inherited breast cancer in the breast oncology care setting. Cancer Control 19, 255–266 (2012).
Stuppia, L. BRCA1 and BRCA2 molecular testing in women with different risk of hereditary breast cancer: cost/effectiveness and psychological implications. Curr. Womens Health Rev. 8, 12–16 (2012).
Halbert, C. H. Decisions and outcomes of genetic testing for inherited breast cancer risk. Ann. Oncol. 15, I35–I39 (2004).
Cipollini, G., Tommasi, S., Paradiso, A., Aretini, P., Bonatti, F., Brunetti, I. et al. Genetic alterations in hereditary breast cancer. Ann. Oncol. 15, I7–I13 (2004).
Barcenas, C. H., Hosain, G. M., Arun, B., Zong, J., Zhou, X., Chen, J. et al. Assessing BRCA carrier probabilities in extended families. J. Clin. Oncol. 24, 354–360 (2006).
Marroni, F., Aretini, P., D'Andrea, E., Caligo, M. A., Cortesi, L., Viel, A. et al. Evaluation of widely used models for predicting BRCA1 and BRCA2 mutations. J. Med. Genet. 41, 278–285 (2004).
Ready, K. J., Vogel, K. J., Atchley, D. P., Broglio, K. R., Solomon, K. K., Amos, C. et al. Accuracy of the BRCAPRO model among women with bilateral breast cancer. Cancer 115, 725–730 (2009).
Bodmer, D., Ligtenberg, M. J., van der Hout, A. H., Gloudemans, S., Ansink, K., Oosterwijk, J. C. et al. Optimal selection for BRCA1 and BRCA2 mutation testing using a combination of 'easy to apply' probability models. Br. J. Cancer 95, 757–762 (2006).
Acknowledgements
We thank Drs Patrizia Di Fulvio, Paola Scioletti and Stefano Valbonesi for technical support. This work was supported by a grant of the G. d’Annunzio University to LS.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Antonucci, I., Provenzano, M., Sorino, L. et al. Comparison between CaGene 5.1 and 6.0 for BRCA1/2 mutation prediction: a retrospective study of 150 BRCA1/2 genetic tests in 517 families with breast/ovarian cancer. J Hum Genet 62, 379–387 (2017). https://doi.org/10.1038/jhg.2016.138
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2016.138