Abstract
Developmental dyslexia (DD) is a neurodevelopment disorder characterized by reading disabilities without apparent etiologies. Nonsyndromic cleft lip with or without cleft palate (NSCL/P) is a structural craniofacial malformation featured by isolated orofacial abnormalities. Despite substantial phenotypic differences, potential linkage between these two disorders has been suggested as prevalence of DD among NSCL/P patients was much higher than that in general populations. Previous neuroimaging studies observed impaired short-term memory in patients with DD and NSCL/P, respectively. Genetic factors have a fundamental role during neurodevelopment and craniofacial morphogenesis but there lacks of evidence to support the linkage between DD and NSCL/P at genetic level. A recent genome-wide association study in Chinese populations identified a number of genetic polymorphisms associated with NSCL/P. Herein, we selected three risk variants of NSCL/P namely rs8049367, rs4791774 and rs2235371, and performed association analysis with DD in a Chinese population consisting 631 elementary school-aged children with 288 dyslexic cases without NSCL/P and 343 healthy controls. After Bonferroni correction for multiple comparisons, the T allele of rs8049367 showed significant association with DD (OR=1.41, P=0.0085). It is an intergenic variant between CREBBP and ADCY9 located at 16p13.3. The CREBBP gene was reported to have an essential role during memory formation, although ADCY9 was involved in dental development. In future studies, understanding functional effects of rs8049367 on CERBBP and ADCY9 might contribute to explain underlying etiologies shared by DD and NSCL/P.
Similar content being viewed by others
Introduction
Developmental dyslexia (DD), also known as reading disability, is characterized by unexpected difficulties in reading and spelling without apparent intellectual and neurological impairment.1 The prevalence of reading disability among children and adolescents ranges from 5 to 17.5%.2 Notably, it was more prevalent among children with nonsyndromic cleft lip with or without cleft palate (NSCL/P), a common birth defect characterized by isolated orofacial cleft with unknown but complex etiology.3 The rate of moderate and severe degree of reading disability were reported to be 35 and 17%, respectively, in a group of NSCL/P children.4
Although DD and NSCL/P are apparently different, they might have potential common etiologic pathways. Compared with age-matched non cleft controls, children with NSCL/P scored significantly lower in a series of reading measurements.5, 6 It has been suggested that DD and NSCL/P shared similar neurological endophenotypes. Short-term verbal memory is the ability to keep verbal information in conscious awareness for brief periods of time.7 Deficiency in short-term verbal memory was known to be an important endophenotype of DD.7 Among children with NSCL/P, reading disability were also featured by short-term verbal memory defects which fitted into a classic model of DD.8, 9 Therefore, there might be some common etiologies underlying DD and NSCL/P. Neuroimaging studies indicated a neurological basis of DD could be explained by aberrant structure and function of reading and language networks throughout the left hemisphere.10 Consistent with this view, the most severely affected brain region among NSCL/P patients was reported to be the left temporal lobe in which observed structural abnormalities were directly related to cognitive dysfunctions.11
In addition to neurological findings, DD and NSCL/P might have similar etiologies at genetic level as well. Both DD and NSCL/P are complex developmental disorders with genetic and non-genetic components to their perspective etiologies. Current genetic understanding of DD is mainly based on linkage analysis and candidate gene association study. Linkage studies of DD identified nine DD susceptibility loci termed DYX1 to DYX9. Further refinement of these loci reported a number of candidate genes associated with DD. The well recognized candidate genes of DD includes DYX1C1 in DYX1, DCDC2 and KIAA0319 in DYX2 and ROBO1 in DYX5.12, 13, 14, 15 In recent years, some of these associated genes have been replicated in different populations through association studies.16 Meanwhile, genetic research of NSCL/P has recently identified multiple genes associated with disease susceptibility through genome-wide association study (GWAS). Previous GWAS in western populations identified a number of representative single nucleotide polymorphisms (SNPs) strongly identified as NSCL/P susceptibility loci.17, 18, 19, 20 Given substantial differences in genetic backgrounds, their exact contribution to risk in Chinese populations remains unknown. Recently, the first GWAS of NSCL/P in Chinese reported several risk SNPs of NSCL/P.21 In particular, rs8049367 located in 16p13.3 was found to be a risk variant exclusive to Chinese.21 We also confirmed variants within previously reported susceptibility loci including rs4791774 located in 17p13.1 and rs2235371 located in 1q32.2.21
Given the substantial genetic components underlying DD and NSCL/P, we aimed to explore observed linkage between these two disorders from genetic perspective. In the present study, we selected NSCL/P susceptibility SNPs from our recent GWAS report and evaluated their contribution to DD in a Chinese case–control study. Our study provided additional evidences to justify underlying genetic etiologies shared by DD and NSCL/P.
Materials and methods
Subjects
The criteria for participants with DD and healthy individuals were described elsewhere previously.22 Briefly, DD screening underwent the two-stage procedures. First, primary school students aged between 7 and 13 from Shandong province of China were subjected to a Chinese reading test consisting of character-, word- and sentence-level questions. The participants whose reading scores were above 87th percentile or below the 13th percentile among all students in the same grade were chosen for further evaluation. These participants were subjected to a character reading test composed of 300 Chinese characters individually for the assessment of reading ability. Then the Raven’s standard test was performed to exclude individuals with intelligence deficiency. Finally, a total of 631 children, 288 dyslexic participants and 343 controls, were selected for subsequent analysis. This study was approved by the ethical committee of Tsinghua University School of Medicine. All participants were informed with written consent.
Candidate SNPs genotyping
Three SNPs reported to be strongly associated with NSCL/P, rs2235371, rs8049367 and rs4791774, were genotyped on Sequenom MassARRAY platform (Sequenom, San Diego, CA, USA) at CapitalBio Corporation (Beijing, China). Genomic DNA samples were extracted from saliva samples using Oragene DNA self-collection kit (DNA Genotek, Ottawa, Ontario, Canada) and DNA quantity was determined by Nanodrop spectrophotometry (Nanodrop 1000 Spectrophotometer, Thermo Scientific, Wilmington, DE, USA). A PCR reaction based on a locus-specific primer extension reaction was designed using the MassARRAY Assay Design software package (v3.1, Sequenom, San Diego, CA, USA). MALDI-TOF mass spectrometer (Sequenom) and MassARRAY Type 4.0 software (Sequenom) were used for mass determination and data acquisition.
Data analysis
Statistical analysis was undertaken using PLINK software version 1.9 (http://pngu.mgh.harvard.edu/~purcell/plink/), which is an open-source whole-genome association analysis toolset and is commonly used to perform a range of basic, large-scale analyses.23 Hardy–Weinberg equilibrium tests were undertaken for each SNP, and association tests were performed using additive, dominant or recessive genetic models under a logistic regression model with covariates of age and sex. Bonferroni corrections for multiple testing were used to determine statistical significance.
Results
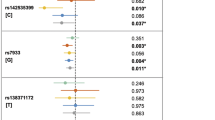
Basic characteristics of research participants are presented in Table 1. There were no differences of age distribution between DD cases and controls. The proportion of male in DD case group was higher than control groups. The effects of gender on DD were indicated in Supplementary Table S1. Three SNPs were genotyped in current study. In our sample, the allele T of rs2235371 was less frequent in cases than that in controls. The allele T of rs8049367 and the allele G of rs4791774 were more frequent in cases than that in controls.
SNP rs2235371 was not significantly associated with DD under allelic (P=0.1905, odd ratio (OR)=0.8569), additive (P=0.1780, OR=0.8487) and recessive (P=0.5807, OR=1.1470) models, except dominant model (P=0.0293, OR=0.6998). However, with the adjustment of age and sex, the association under dominant model became nonsignificant (P=0.1659). SNP rs8049367 was significantly associated with DD under additive (P=0.0085, OR=1.4100), dominant (P=0.0256, OR=1.4960) and recessive (P=0.0374, OR=1.7280) model, after adjustment for age and sex. SNP rs4791774 was not significantly associated with DD under all models (Table 2).
After the Bonferonni correction for multiple comparisons, only SNP rs8049367 (OR=1.4100, 95% CI=1.0920–1.8220) on chromosome 17 was significantly associated with DD under additive models, indicating rs8049367 is a potential SNP marker for DD.
Discussion
In total, three susceptibility SNPs of NSCL/P were evaluated for their contribution to risk to DD. These SNPs were initially reported in a recent GWAS report as novel risk variants for NSCL/P specific to Chinese populations.21 In the present study, the T allele of rs8049367 showed 41% increased risk of DD, therefore rs8049367 appears to be susceptibility SNP for DD as well. However, the observed association between rs8049367 and DD should be interpreted with caution as this SNP is an intergenic variant located 50 kb upstream of CREBBP and 32 kb downstream of ADCY9 with unknown function. The CREBBP gene encodes a nuclear protein that binds to cAMP response element binding protein (CREB) and acts a transcriptional coactivator with histone acetyltransferase activity.24, 25 Mutations in CREBBP gene are known to cause Rubinstein–Taybi syndrome, which is a genetic disorder characterized by developmental delay and abnormalities as well as intellectual disabilities.26 In addition, the CREBBP gene was implicated in memory consolidation.27, 28 Knock-out of CREBBP in mice resulted in permanent impairments on both long- and short-term memory.29 Meanwhile, upregulation of CREBBP-mediated transcriptional activity improved both long- and short-term memory.30 Short-term verbal memory has a critical role in phonological processing, comprehension, and other reading-related processes. Previous studies of DD frequently observed impaired short-term verbal memory among dyslexics. In a recent study, Chinese dyslexics showed reduced short-term verbal memory during recall of Chinese characters than age-matched normal readers.31 Given reported implication of CREBBP in memory impairment as well as developmental and intellectual abnormalities, understanding functional effects of rs8049367 on CREBBP in future studies might explain its contribution to DD.
There were considerable amounts of NSCL/P patients showed reduced reading abilities.4 Previous observed reading disabilities among NSCL/P were also featured by impaired short-term verbal memory which suggested underlying etiologies shared by DD and NSCL/P.8 As rs8049367 has been associated with NSCL/P in the recent GWAS, our preliminary results indicated rs8049367 as a shared risk factor for both disorders. During palate development, expression of CREBBP was constant. The CREBBP-mediated transcription is likely regulated by changes in its phosphorylation rather than by changes in its expression.32 Meanwhile, expression of ADCY9 was upregulated in the dental pulp stem cultures of NSCL/P patients.21 Therefore, rs8049367 might contribute to DD and NSCL/P through its pleiotropic effects on CREBBP and ADCY9, respectively. Indeed, rs8049367 was in strong LD with an exon variant of a long non-coding RNA (lncRNA), namely RP11–462G12.2, which was predicted to interact with both CREBBP and ADCY9.21 Thus, future investigation on lncRNA RP11–462G12.2 might contribute to explain the assumed pleiotropic effects of rs8049367.
In conclusion, we performed association study of three NSCL/P susceptibility SNPs with DD in a Chinese population and observed significant association of rs8049367. This intergenic variant might have pleiotropic effects on its two nearby genes CERBBP and ADCY9. Further functional investigations are warranted to validate the assumed pleiotropic effects which might explain the shared etiology underlying DD and NSCL/P.
References
Paracchini, S., Scerri, T. & Monaco, A. P. The genetic lexicon of dyslexia. Annu. Rev. Genomics Hum. Genet. 8, 57–79 (2007).
Shaywitz, S. E. Dyslexia. N. Engl. J. Med. 338, 307–312 (1998).
Mossey, P. A., Little, J., Munger, R. G., Dixon, M. J. & Shaw, W. C. Cleft lip and palate. Lancet. 374, 1773–1785 (2009).
Richman, L. C., Eliason, M. J. & Lindgren, S. D. Reading disability in children with clefts. Cleft Palate J. 25, 21–25 (1988).
Collett, B. R., Stott-Miller, M., Kapp-Simon, K. A., Cunningham, M. L. & Speltz, M. L. Reading in children with orofacial clefts versus controls. J. Pediatr. Psychol. 35, 199–208 (2010).
Conrad, A. L., McCoy, T. E., DeVolder, I., Richman, L. C. & Nopoulos, P. Reading in subjects with an oral cleft: speech, hearing and neuropsychological skills. Neuropsychology 28, 415–422 (2014).
Peterson, R. L. & Pennington, B. F. Developmental dyslexia. Lancet 379, 1997–2007 (2012).
Richman, L. C. & Ryan, S. M. Do the reading disabilities of children with cleft fit into current models of developmental dyslexia? Cleft Palate Craniofac. J. 40, 154–157 (2009).
Richman, L. C., Wilgenbusch, T. & Hall, T. Spontaneous verbal labeling: visual memory and reading ability in children with cleft. Cleft Palate Craniofac J. 42, 565–569 (2005).
Peterson, R. L. & Pennington, B. F. Developmental dyslexia. Annu. Rev. Clin. Psychol. 11, 283–307 (2015).
Nopoulos, P., Berg, S., Canady, J., Richman, L., Van Demark, D. & Andreasen, N. C. Structural brain abnormalities in adult males with clefts of the lip and/or palate. Genet. Med. 4, 1–9 (2002).
Taipale, M., Kaminen, N., Nopola-Hemmi, J., Haltia, T., Myllyluoma, B., Lyytinen, H. et al. A candidate gene for developmental dyslexia encodes a nuclear tetratricopeptide repeat domain protein dynamically regulated in brain. Proc. Natl Acad. Sci. USA 100, 11553–11558 (2003).
Meng, H., Smith, S. D., Hager, K., Held, M., Liu, J., Olson, R. K. et al. DCDC2 is associated with reading disability and modulates neuronal development in the brain. Proc. Natl Acad. Sci. USA 102, 17053–17058 (2005).
Cope, N., Harold, D., Hill, G., Moskvina, V., Stevenson, J., Holmans, P. et al. Strong evidence that KIAA0319 on chromosome 6p is a susceptibility gene for developmental dyslexia. Am. J. Hum. Genet. 76, 581–591 (2005).
Hannula-Jouppi, K., Kaminen-Ahola, N., Taipale, M., Eklund, R., Nopola-Hemmi, J., Kääriäinen, H. et al. The axon guidance receptor gene ROBO1 is a candidate gene for developmental dyslexia. PLoS Genet. 1, e50 (2005).
Sun, Y., Gao, Y., Zhou, Y., Chen, H., Wang, G., Xu, J. et al. Association study of developmental dyslexia candidate genes DCDC2 and KIAA0319 in Chinese population. Am. J. Med. Genet. B Neuropsychiatr. Genet. 165B, 627–634 (2014).
Birnbaum, S., Ludwig, K. U., Reutter, H., Herms, S., Steffens, M., Rubini, M. et al. Key susceptibility locus for nonsyndromic cleft lip with or without cleft palate on chromosome 8q24. Nat. Genet. 41, 473–477 (2009).
Beaty, T. H., Murray, J. C., Marazita, M. L., Munger, R. G., Ruczinski, I., Hetmanski, J. B. et al. A genome-wide association study of cleft lip with and without cleft palate identifies risk variants near MAFB and ABCA4. Nat. Genet. 42, 525–529 (2010).
Mangold, E., Ludwig, K. U., Birnbaum, S., Baluardo, C., Ferrian, M., Herms, S. et al. Genome-wide association study identifies two susceptibility loci for nonsyndromic cleft lip with or without cleft palate. Nat. Genet. 42, 24–26 (2010).
Ludwig, K. U., Mangold, E., Herms, S., Nowak, S., Reutter, H., Paul, A. et al. Genome-wide meta-analyses of nonsyndromic cleft lip with or without cleft palate identify six new risk loci. Nat. Genet. 44, 968–971 (2012).
Sun, Y., Huang, Y., Yin, A., Pan, Y., Wang, Y., Wang, C. et al. Genome-wide association study identifies a new susceptibility locus for cleft lip with or without a cleft palate. Nat. Commun 6, 6414 (2015).
Tan, L. H., Xu, M., Chang, C. Q. & Siok, W. T. China’s language input system in the digital age affects children’s reading development. Proc. Natl Acad. Sci. USA 110, 1119–1123 (2013).
Purcell, S., Neale, B., Todd-Brown, K., Thomas, L., Ferreira, M. A. R., Bender, D. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Reddy, C. S. Alterations in protein kinase A signalling and cleft palate: a review. Hum. Exp. Toxicol. 24, 235–242 (2005).
Goodman, R. H. & Smolik, S. CBP/p300 in cell growth, transformation, and development. Genes Dev. 14, 1553–1577 (2000).
Petrij, F., Giles, R. H., Dauwerse, H. G., Saris, J. J., Hennekam, R. C., Masuno, M. et al. Rubinstein-Taybi syndrome caused by mutations in the transcriptional co-activator CBP. Nature 376, 348–351 (1995).
Korzus, E., Rosenfeld, M. G. & Mayford, M. CBP histone acetyltransferase activity is a critical component of memory consolidation. Neuron 42, 961–972 (2004).
Vecsey, C. G., Hawk, J. D., Lattal, K. M., Stein, J. M., Fabian, S. A., Attner, M. A. et al. Histone deacetylase inhibitors enhance memory and synaptic plasticity via CREB:CBP-dependent transcriptional activation. J. Neurosci. 27, 6128–6140 (2007).
Chen, G., Zou, X., Watanabe, H., van Deursen, J. M. & Shen, J. CREB binding protein is required for both short-term and long-term memory formation. J. Neurosci. 30, 13066–13077 (2010).
Suzuki, A., Fukushima, H., Mukawa, T., Toyoda, H., Wu, L.-J., Zhao, M.-G. et al. Upregulation of CREB-mediated transcription enhances both short- and long-term memory. J. Neurosci. 31, 8786–8802 (2011).
Zhao, J., Yang, Y., Song, Y.-W. & Bi, H.-Y. Verbal short-term memory deficits in chinese children with dyslexia may not be a problem with the activation of phonological representations. Dyslexia 21, 304–322 (2015).
Weston, W. M. & Greene, R. M. Developmental changes in phosphorylation of the transcription factor CREB in the embryonic murine palate. J. Cell Physiol. 164, 277–285 (1995).
Acknowledgements
This work was funded by the National Key Basic Research Program Grant of the People’s Republic of China (2012CB720703) and Shenzhen Peacock Plan (Grant No. KQTD2015033016104926). We thank all of the study subjects, research staff and students who participated in this study.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Journal of Human Genetics website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Wang, B., Zhou, Y., Leng, S. et al. Genetic polymorphism of nonsyndromic cleft lip with or without cleft palate is associated with developmental dyslexia in Chinese school-aged populations. J Hum Genet 62, 265–268 (2017). https://doi.org/10.1038/jhg.2016.121
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2016.121
This article is cited by
-
The genetic factors contributing to the risk of cleft lip-cleft palate and their clinical utility
Oral and Maxillofacial Surgery (2022)