Abstract
Short stature homeobox gene (SHOX) is located in the pseudoautosomal region 1 of the sex chromosomes. It encodes a transcription factor implicated in the skeletal growth. Point mutations, deletions or duplications of SHOX or its transcriptional regulatory elements are associated with two skeletal dysplasias, Léri–Weill dyschondrosteosis (LWD) and Langer mesomelic dysplasia (LMD), as well as in a small proportion of idiopathic short stature (ISS) individuals. We have identified a total of 15 partial SHOX deletions and 13 partial SHOX duplications in LWD, LMD and ISS patients referred for routine SHOX diagnostics during a 10 year period (2004–2014). Subsequently, we characterized these alterations using MLPA (multiplex ligation-dependent probe amplification assay), fine-tiling array CGH (comparative genomic hybridation) and breakpoint PCR. Nearly half of the alterations have a distal or proximal breakpoint in intron 3. Evaluation of our data and that in the literature reveals that although partial deletions and duplications only account for a small fraction of SHOX alterations, intron 3 appears to be a breakpoint hotspot, with alterations arising by non-allelic homologous recombination, non-homologous end joining or other complex mechanisms.
Similar content being viewed by others
Introduction
Short stature homeobox gene (SHOX, MIM 312865) is located in the pseudoautosomal region 1 (PAR1) of the sex chromosomes.1, 2 It encodes a transcriptional factor implicated in the skeletal growth.3 SHOX is regulated by two promoters, located in exon 1 and the non-coding part of exon 2 and seven transcriptional regulatory elements (three upstream and four downstream of SHOX).4, 5, 6, 7, 8
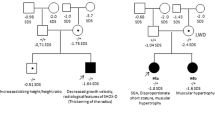
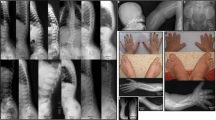
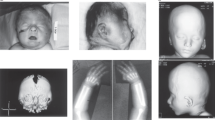
Alterations in SHOX are associated with two skeletal dysplasias, Léri–Weill dyschondrosteosis (LWD, MIM 127300)9, 10 and Langer mesomelic dysplasia (LMD, MIM 249700)10 and a small proportion of idiopathic short stature cases (ISS, MIM 300582), classified as individuals with a height below −2 standard deviations (SDS) in the absence of known specific causative disorders.11 LWD is a dominantly inherited skeletal dysplasia characterized by disproportionate short stature, mesomelic limb shortening and the characteristic Madelung deformity of the forearm: the bowing of the radius, the distal dislocation of the ulna and triangulation of the carpal bones. To date, heterozygous deletions and duplications of three distinct regions of the PAR1, SHOX and the two enhancer region intervals, located upstream and downstream of SHOX or mutations within SHOX have been identified in ~60–80% of LWD and 2–5% of ISS cases.12, 13, 14 LMD is a more severe skeletal dysplasia due to homozygous or compound heterozygous mutations in SHOX or its enhancers.15, 16, 17, 18
Recently, we demonstrated that the cis-regulatory region of SHOX extends to ~1 Mb of the PAR1,19 which suggests that further SHOX regulatory elements may be present and that alterations of these regions may be the cause of the phenotype in LWD, LMD and ISS patients. This is supported by the observation of deletions located further downstream from the known SHOX enhancer interval in two patients with an intermediate phenotype between LWD and LMD.20, 21
Here we report the identification and characterization of 15 novel partial SHOX deletions and 13 partial duplications, detected during routine genetic analysis and review these and other intragenic deletions and duplications described in the literature.
Materials and Methods
Clinical patients
The LWD, LMD and ISS patient samples were recruited from endocrinology and genetic clinics over a 10 year period (2004–2014). Genomic DNA was isolated from peripheral blood using the Blood kit (QIAGEN, Valencia, CA, USA) or Chemagic DNA extraction special kit (Chemagen, Perkin Elmer, Baesweiler, Germany). Suspected LWD patients were ascertained using the inclusion criteria of bilateral Madelung deformity and mesomelic shortening of the limbs in the proband or a direct family member. ISS patients with stature <−2 SDS were ascertained using the current consensus criteria.11 The study was approved by the local ethical committees and all participants provided informed consent.
SHOX/PAR1 deletion detection
Analysis of SHOX/PAR1 was carried out by a combination of MLPA (multiplex ligation-dependent probe amplification assay), array CGH (comparative genomic hybridation), fluorescent in situ hybridization (FISH) and microsatellite analysis as previously described.22 MLPA analysis was carried out using the commercial SHOX and PAR1 MLPA kits (Salsas P018B/C/D1/E1/F1/G1, MRC Holland, Amsterdam, The Netherlands). In particular cases, the deletions were delimited using custom-designed MLPA assays, one for the upstream region of SHOX23 and another including sequences flanking SHOX exon 6a (Supplementary Table 1). Data was analyzed as described previously.23
Array CGH
Y-chromosome-specific file-tiling array CGH (Roche NimbleGen Systems, Madison, WI, USA) were hybridized and analyzed using the service provided by ImaGenes, Berlin, Germany. Data was analyzed and visualized with Signalmap software (NimbleGen Systems). For each spot on the array, log2-ratios of the Cy3-labeled test sample versus the Cy5-labeled reference sample were calculated.
Deletion breakpoint determination
Oligonucleotides were designed in regions shown to be present as two copies by aCGH or custom-designed MLPA. The oligos for crossing the deletion breakpoint identified in proband 9 were 5′-GCTAAACTGCCTGCACTCTC-3′ and 5′-ACCAGAAGCTCCAGCGTCTT-3′, probands 13 and 14: 5′-GGTGGAAACTTCGGTTCTC-3′ and 5′-TGCGCCCTTCTTAACCAG-3′, and for proband 15, 5′-CTTCTTGTACCGTCTTTTGCC-3′ and 5′-AAATCAAACTGAAACCGTCCC-3′. Amplified products were directly sequenced with the amplification primers, with the exception of the breakpoint PCR of proband 15 where an internal sequence primer 5′-GAACCTACTTCCCAAAGATTC-3′ was required, or cloned into a pCR2.1 vector using the TA cloning kit (Invitrogen, Carlsbad, CA, USA) and subsequently sequenced (proband 9). The breakpoints were determined using Sequencher V5.1 (Gene Codes Corporation, Ann Arbor, MI, USA) and Ensembl (www.ensembl.org).
The deletion mechanism was subsequently investigated by computational analysis. Homology and repeat analyses of the breakpoint flanking sequences (2 kb) was assessed using LALIGN (http://www.ch.embnet.org/software/LALIGN_form.html) and RepeatMasker (http://www.repeatmasker.org), respectively. The percentage identity for Repeat Elements observed at the breakpoints was calculated using the Align Sequences Nucleotide BLAST (bl2Seq, http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Results
Partial SHOX deletions were identified using the commercial MLPA, in 15 patients, 13 diagnosed with LWD and 2 with LMD (probands 5 and 9), during the 10 year study period (Figure 1). Six deletions extended from the 5′ region of SHOX and included various exons (probands 1, 2, 3, 4, 5 and 6), eight were intragenic deletions (probands 7, 8, 10, 11, 12, 13, 14 and 15) and one deletion extended from SHOX exon 4 to the downstream region (proband 9; Figure 1). One of the intragenic deletions was a deletion of a single exon, exon 6a (proband 15). Initially we excluded the presence of a polymorphism in the oligonucleotide sequence resulting in allele drop out. Subsequently, we delimited the deletion using a custom-designed MLPA and crossed the junction breakpoint. In addition, patients 10 and 13 had another PAR1 deletion, a deletion of the L05101 probe, which we had demonstrated to correspond to a ~4.9 kb non-pathogenic downstream deletion (Figure 1).24
Schematic representation of the 15 partial SHOX (short stature homeobox gene) deletions detected by MLPA (multiplex ligation-dependent probe amplification assay) SHOX/PAR1 (pseudoautosomal region 1) salsa P018 B1/C1/D1/E1/F1/G1 and the custom-designed MLPAs. Gray boxes indicate the location of the P018-D1 MLPA probes. ▴ Indicate deleted probes. The deletion size range, determined by MLPA data, is indicated adjacent to each deletion. Diagram of region using X chromosome assembly (GRCh37/hg19) but not drawn to scale. This deletion of probe L5101 is due to a non-pathogenic ~4.9 kb deletion.24 Probe L5101 was eliminated from the commercial MLPA from P018E1 onwards.
Seven of the deletions were further delimited using Y-chromosome fine-tiling aCGH (probands 1, 6, 7, 9, 10, 11 and 13; Figure 2; Supplementary Table 2). Further characterization of the remaining patients was not possible due to lack of DNA. Interestingly, aCGH revealed that patient 10 had three PAR1 deletions, one of the deletions previously detected by MLPA was actually composed of two deletions separated by a region of normal dosage (Figure 2; Supplementary Figure 1). Analysis of parental samples revealed that the SHOX deletion and the ~11.7 kb downstream deletion were inherited on the paternal allele whereas the non-pathogenic downstream deletion of ~4.9 kb was maternally inherited. Patients 7, 11 and 13 also had this non-pathogenic deletion.
Array CGH (comparative genomic hybridation) of seven probands with partial SHOX (short stature homeobox gene) deletions. Y-chromosome specific fine-tiling aCGH (NimbleGen) was utilized with a dye-swap experiment. Data from one array of each analyzed proband is shown using the SignalMap software (NimbleGen). No aCGH was undertaken in the remainder of the proband due to lack of DNA. The partial SHOX deletions are indicated with a red asterisk and the non-pathogenic 4959 bp downstream deletion with a blue asterisk. Y-chromosome sequence coordinates are taken from GRCh36/hg18. A full color version of this figure is available at the Journal of Human Genetics journal online.
Partial SHOX duplications were identified in 13 LWD and ISS patients, 10 of which were previously described (Supplementary Figure 3).14 Five partial duplications extended from 5′of SHOX, four of which extended to exon 3 (probands 17–20), seven were intragenic duplications (probands 21–27) and one duplication extended from exon 4 to ~150 kb downstream of SHOX (patient 28).
Seven deletions (~48%, probands 8–14) and five duplications (~38.5%, probands 17–20 and 28) had a distal or proximal breakpoint in intron 3, followed by five deletions (~33%; 5/15; probands 1–4 and 6) and one duplication (proband 21) in intron 2, whereas the remaining probands had breakpoints in distinct regions (Figure 1; Supplementary Figure 3).
The deletion breakpoints were identified in patients 9, 13, 14 and 15, which allowed the precise determination of the deletion extensions: 261 684, 1344, 1681 and 2949 bp, respectively, three of which were located in intron 3 (probands 9, 13 and 14; Figure 3). No homology was observed at the breakpoint flanking regions in patient 9. However, repetitive elements, type Alu were identified at both breakpoints of patients 13 and 14, AluSz/AluSx (78% homology) and AluSz/AluSx (79% homology), respectively. Microhomology of 37, 19 and 7 bp was observed at the 5′and 3′ breakpoints in patients 13, 14 and 15, respectively (Figure 3).
Breakpoint characterization of the partial SHOX (short stature homeobox gene) deletions of individuals 9, 13, 14 and 15. The vertical bar indicates the fusion points of the intragenic deletions and gray rectangle indicates the sequence of unknown origin in the proband 9 and the overlap sequence identical between distal and proximal breakpoint sequence in probands 13, 14 and 15. The exact chromosomal locations are indicated above the vertical bars. PAR1 (pseudoautosomal region 1) sequence coordinates are taken from the X chromosome assembly (GRCh37/hg19). The same oligonucleotides were used for crossing the deletion breakpoints in probands 13 and 14, despite the unavailability of aCGH (array comparative genomic hybridation) data for proband 14. A full color version of this figure is available at the Journal of Human Genetics journal online.
Discussion
For many years, SHOX genetic diagnosis in LWD, LMD and ISS patients was undertaken by microsatellite analysis and/or FISH. During the past decade, MLPA, qPCR (quantitative PCR) and aCGH have considerably improved the detection of SHOX alterations. MLPA and direct sequencing remain the general practice as next-generation sequencing has not replaced these techniques as SHOX and its enhancers are poorly captured.
During the 10-year study period, we have identified and characterized 15 different partial SHOX deletions. All deletion extensions were different and sizes ranged from 1344 to ~374 kb. Exact breakpoints were characterized in four individuals, probands, 9, 13, 14 and 15.
Proband 9, diagnosed with LMD, had a partial SHOX deletion in one allele and a frameshift mutation in exon 6 in the other allele, c.722_723insC.16 The deletion extended from intron 3 and included downstream sequence. Although no homologous repeat was observed at the 5′ breakpoint, an AluSz sequence was present at the 3′ breakpoint. No microhomology was present at the breakpoints but 50 bases had been incorporated at the deletion junction, 42 of which constitute part of the interspersed repeat sub-family, endogenous retrovirus group K (ERVK). Although long terminal repeats are known to give arise to deletions and duplications through homologous recombination, we do not fully understand how this sequence inserted into the deletion region.
Both probands 13 and 14 had an intragenic deletion including exons 4 and 5. The deletion breakpoints in introns 3 and 5 were different, thus leading to different deletion sizes of 1344 and 1681 bp, respectively. Alu elements were identified at both breakpoints, AluSz/AluSx (78% homology) and AluSz/AluSx (79% homology) and with an overlap sequence of 37 and 19 nucleotides respectively, therefore suggesting non-allelic homologous recombination event as the most probable deletion mechanism.
A rare single exon deletion was observed in proband 15, which extended from SHOX intron 5 to intron 6a. No repetitive elements were observed at the breakpoints, but a homologous sequence of seven nucleotides was found, thus, suggesting that it had occurred by non-homologous end joining mechanism.
Although the breakpoints were not determined in proband 10, aCGH data showed that one of the MLPA detected deletions was actually composed of two deletions separated by a region of normal dosage. Thus, proband 10 had three deletions: a SHOX deletion and two downstream PAR1 deletions. Parental analysis showed that the SHOX deletion and the deletion in the 3′end of SHOX were paternally inherited whereas the non-pathogenic ~4959 bp downstream PAR1 deletion was present on the maternal allele.24 The identification of two deletions on the paternal allele suggests a different deletion mechanism. The breakpoints have not been finely characterized but we hypothesize that it may have arisen by a replicative mechanism such as FoSTeS (fork stalling and template switching),25 SRS (serial replication slippage) or MMBIR (microhomology-mediated break induced replication).26
A total of 21 partial SHOX deletions and 38 partial SHOX duplications have been reported in the literature,27, 28, 29, 30, 31, 32, 33, 34, 35, 36 13 (62%) of the deletions and 23 (60.5%) of the duplications have one of its breakpoints in intron 3.30, 32, 33, 35, 36 The breakpoints have only been characterized in three deletions30 and two duplications.35
The three delimited deletions were shown to include exons 4 and 5 (5906 bp, case 1), exons 4–6a (5594 bp, case 2) and exon 4–6b (50199 bp, case 3).30 Non-repetitive sequences were identified at the breakpoint junctions in cases 1 and 2, whereas Alu elements were present in case 3, thus, non-homologous end joining and non-allelic homologous recombination could explain the deletion mechanisms respectively.30 Also, two single exon 1 deletions have been identified in ISS patients.28, 34 SHOX is transcribed from two alternative promoters, located in exon 1 (P1) and the non-coding part of 2 (P2). The P2 promoter has been shown to be the strongest promoter whereas the P1 promoter controls fine tuning of SHOX expression, by translational regulation.4 Thus, the P1 promoter is deleted in these two individuals, but the main P2 promoter is intact in these two ISS individuals. The pathogenicity of these deletions are unknown but the authors propose that they result in SHOX haploinsufficiency28 or that the deletion causes the deletion of a oligopyrimidine tract and seven AUG codons upstream of the open reading frame, thus affecting translation.34
All partial SHOX deletions have been associated with LWD or short stature, whereas partial SHOX duplications have been identified in patients with variable phenotypes: short stature or LWD,14, 33, 35 Mayer-Rokitansky-Kuster-Hauser Syndrome,32 autism spectrum disorders and related neurodevelopment conditions.36 The variable clinical manifestations associated with partial or complete SHOX duplications are likely to be dependent on the physical localization of the duplicated sequence.14
In contrast to complete deletions of SHOX, where we failed to observe a hotspot region,37 partial SHOX deletion breakpoints seem to occur frequently in intron 3 (7/15 partial deletion). All characterized breakpoints have occurred in an interval of 2599 bp of the 3′ part of intron 3 (Supplementary Figure 2). Intron 3 spans a total of 5994 bp, 28.6% of which consists of repetitive sequences: 13.76% Alu elements (two AluSz and one AluSx1), 14.33% simple repeats (two AT-rich regions) and 0.53% A-rich, but not all the breakpoints occurred within these elements. Indeed it is the largest SHOX intron with a greater number of Alu elements and AT-rich region with respect to the other introns: intron 1 (62.87% simple repeats), intron 2 (9% repeats elements and 0.64% simple repeats) and intron 5 (12.90% AluSx). Only three of the five characterized breakpoints, two in this study and one described by Fukami et al.,35 were localized to these repetitive elements.
In our entire LWD, LMD and ISS referral cohort we have observed a smaller number of complete duplications compared with complete SHOX deletions (16 versus 82), and in the case of partial deletions and duplications we have observed approximately the same number of cases (15 versus 13),14 but no reciprocal deletion and duplication have been observed, as might have been expected and observed in other disorders.38, 39, 40 Five of the 13 (38.5%) duplication breakpoints were located in intron 3 but once again, no specific sequences were implicated. Thus, a total of 12/28 (42.8%) of the deletions and duplications occurred within this intron.
In summary, we have identified 15 partial SHOX deletions and 13 partial SHOX duplications in LWD and LMD cases during the last decade. Approximately 50% of the breakpoints are located in intron 3, which may be a region with higher predisposition to breakage although the reason why remains unclear.
References
Ellison, J. W., Wardak, Z., Young, M. F., Gehron-Robey, P., Laig-Webster, M. & Chiong, W. PHOG, a candidate gene for involvement in the short stature of Turner syndrome. Hum. Mol. Genet. 6, 1341–1347 (1997).
Rao, E., Weiss, B., Fukami, M., Rump, A., Niesler, B., Mertz, A. et al. Pseudoautosomal deletions encompassing a novel homeobox gene cause growth failure in idiopathic short stature and Turner syndrome. Nat. Genet. 16, 54–63 (1997).
Rao, E., Blaschke, R. J., Marchini, A., Niesler, B., Burnett, M. & Rappold, G. A. The Leri-Weill and Turner syndrome homeobox gene SHOX encodes a cell-type specific transcriptional activator. Hum. Mol. Genet. 10, 3083–3091 (2001).
Blaschke, R. J., Topfer, C., Marchini, A., Steinbeisser, H., Jansen, J. W. G. & Rappold, G. U. Transcriptional and translational regulation of the Léri-Weill and Turner syndrome homeobox gene SHOX. J. Biol. Chem. 278, 47839–47826 (2003).
Fukami, M., Okuyama, T., Yamamori, S., Nishimura, G. & Ogata, T. Microdeletion in the SHOX 3' region associated with skeletal phenotypes of Langer mesomelic dysplasia in a 45,X/46,X,r(X) infant and Léri-Weill dyschondrosteosis in her 46,XX mother: implication for the SHOX enhancer. Am. J. Med. Genet. A 137, 72–76 (2005).
Sabherwal, N., Bangs, F., Roth, R., Weiss, B., Jantz, K., Tiecke, E. et al. Long-range conserved non-coding SHOX sequences regulated expression in developing chicken limb and are associated with short stature phenotypes in human patients. Hum. Mol. Genet. 16, 210–222 (2007).
Durand, C., Bangs, F., Signolet, J., Decker, E., Tickle, C. & Rappold, G. Enhancer elements upstream of the SHOX gene are active in the developing limb. Eur. J. Hum. Genet. 18, 527–532 (2009).
Benito-Sanz, S., Royo, J. L., Barroso, E., Paumard-Hernández, B., Barreda-Bonis, A. C., Liu, P. et al. Identification of the first recurrent PAR1 deletion in Léri-Weill dyschondrosteosis and idiopathic short stature reveals the presence of a novel SHOX enhancer. J. Med. Genet. 49, 442–450 (2012).
Shears, D. J., Vassal, H. J., Goodman, F. R., Palmer, R. W., Reardon, W., Superti-Furga, A. et al. Mutation and deletion of the pseudoautosomal gene SHOX cause Léri-Weill dyschondrosteosis. Nat. Genet. 19, 70–73 (1998).
Belin, V., Cusin, V., Viot, G., Girlich, D., Toutain, A., Moncla, A. et al. SHOX mutations in dyschondrosteosis (Léri-Weill syndrome). Nat. Genet. 19, 67–69 (1998).
Cohen, P., Rogol, A. D., Deal, C. L., Saenger, P., Reiter, E. O., Ross, J. L. et al. Consensus statement on the diagnosis and treatment of children with idiopathic short stature: a summary of the Growth Hormone Research Society, the Lawson Wilkins Pediatric Endocrine Society, and the European Society for Paediatric Endocrinology Workshop. J. Clin. Endocrinol. Metab. 93, 4210–4217 (2007).
Chen, J., Wildhardt, G., Zhong, Z., Röth, R., Weiss, B., Steinberger, D. et al. Enhancer deletions of the SHOX gene as a frequent cause of short stature: the essential role of a 250 kb downstream regulatory domain. J. Med. Genet. 46, 834–839 (2009).
D'haene, B., Hellemans, J., Craen, M., De Schepper, J., Devriendt, K., Fryns, J. P. et al. Improved molecular diagnostics of idiopathic short stature and allied disorders: quantitative polymerase chain reaction-based copy number profiling of SHOX and pseudoautosomal region 1. J. Clin. Endocrinol. Metab. 95, 3010–3018 (2010).
Benito-Sanz, S., Barroso, E., Heine-Suñer, D., Hisado-Oliva, A., Romanelli, V., Rosell, J. et al. Clinical and molecular evaluation of SHOX/PAR1 duplications in Leri-Weill dyschondrosteosis (LWD) and idiopathic short stature (ISS). J. Clin. Endocrinol. Metab. 96, E404–E412 (2011).
Shears, D. J., Guillen-Navarro, E., Sempere-Miralles, M., Domingo-Jimenez, R., Scambler, P. J. & Winter, R. M. Pseudodominant inheritance of Langer mesomelic dysplasia caused by a SHOX homeobox missense mutation. Am. J. Med. Genet. 110, 153–157 (2002).
Zinn, A. R., Wei, F., Zhang, L., Elder, F. F., Scott, C. I. Jr, Marttila, P. et al. Complete SHOX deficiency causes Langer mesomelic dysplasia. Am. J. Med. Genet. 110, 158–163 (2002).
Campos-Barros, A., Benito-Sanz, S., Ross, J. L., Zinn, A. R. & Heath, K. E. Compound heterozygosity of SHOX-encompassing and downstream PAR1 deletions results in Langer mesomelic dysplasia (LMD). Am. J. Med. Genet. A 143A, 933–938 (2007).
Bertorelli, R., Capone, L., Ambrosetti, F., Garavelli, L., Varriale, L., Mazza, V. et al. The homozygous deletion of the 3' enhancer of the SHOX gene causes Langer mesomelic dysplasia. Clin. Genet. 72, 490–491 (2007).
Verdin, H., Fernández-Miñán, A., Benito-Sanz, S., Janssens, S., Callewaert, B., Waele, K. D. et al. Profiling of conserved non-coding elements upstream of SHOX and functional characterisation of the SHOX cis-regulatory landscape. Sci. Rep. 5, 17667 (2015).
Bunyan, D. J., Taylor, E. J., Maloney, V. K. & Blyth, M. Homozygosity for a novel deletion downstream of the SHOX gene provides evidence for an additional long range regulatory region with a mild phenotypic effect. Am. J. Med. Genet. A 164A 11, 2764–2768 (2014).
Tsuchiya, T., Shibata, M., Numabe, H., Jinno, T., Nakabayashi, K., Nishimura, G. et al. Compound heterozygous deletions in pseudoautosomal region 1 in an infant with mild manifestations of langer mesomelic dysplasia. Am. J. Med. Genet. A 164A, 505–510 (2014).
Benito-Sanz, S., del Blanco, D. G., Aza-Carmona, M., Magano, L. F., Lapunzina, P., Argente, J. et al. PAR1 deletions downstream of SHOX are the most frequent defect in a Spanish cohort of Léri-Weill dyschondrosteosis (LWD) probands. Hum. Mutat. 27, 1062 (2006).
Benito-Sanz, S., Aza-Carmona, M., Rodríguez-Estevez, A., Rica-Etxebarria, I., Gracia, R., Campos-Barros, A. et al. Identification of the first PAR1 deletion encompassing upstream SHOX enhancers in a family with idiopathic short stature. Eur. J. Hum. Genet. 20, 125–127 (2012).
Benito-Sanz, S., Aragones, A., Gracia, R., Campos-Barros, A. & Heath, K. E. A non-pathogenic pseudoautosomal region 1 copy number variant downstream of SHOX. Am. J. Med. Genet. A 155A 4, 935–937 (2011b).
Lee, J. A., Carvalho, C. M. & Lupski, J. R. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell 131, 1235–1247 (2007).
Hastings, P. J., Ira, G. & Lupski, J. R. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet. 5, e1000327 (2009).
Huber, C., Rosilio, M., Munnich, A., Cormier-Daire, V. & French SHOX GeNeSIS Module. High incidence of SHOX anomalies in individuals with short stature. J. Med. Genet. 43, 735–739 (2006).
Tan, Y. M. & Loke, K. Y. Isolated haploinsufficiency of exon 1 of the SHOX gene in a patient with idiopathic short stature. J. Clin. Pathol. 59, 773–774 (2006).
Rappold, G., Blum, W. F., Shavrikova, E. P., Crowe, B. J., Roeth, R., Quigley, C. A. et al. Genotypes and phenotypes in children with short stature: clinical indicators of SHOX haploinsufficiency. J. Med. Genet. 44, 306–313 (2007).
Fukami, M., Dateki, S., Kato, F., Hasegawa, Y., Mochizuki, H., Horikawa, R. et al. Identification and characterization of cryptic SHOX intragenic deletions in three Japanese patients with Léri-Weill dyschondrosteosis. J. Hum. Genet. 53, 454–459 (2008).
Funari, M. F., Jorge, A. A., Souza, S. C., Billerbeck, A. E., Arnhold, I. J., Mendonca, B. B. et al. Usefulness of MLPA in the detection of SHOX deletions. Eur. J. Med. Genet. 53, 234–238 (2010).
Gervasini, C., Grati, F. R., Lalatta, F., Tabano, S., Gentilin, B., Colapietro, P. et al. SHOX duplications found in some cases with type I Mayer-Rokitansky-Kuster-Hauser syndrome. Genet. Med. 12, 634–640 (2010).
Bunyan, D. J., Baker, K. R., Harvey, J. F. & Thomas, N. S. Diagnostic screening identifies a wide range of mutations involving the SHOX gene, including a common 47.5 kb deletion 160 kb downstream with a variable phenotypic effect. Am. J. Med. Genet. A 161A, 1329–1338 (2013).
Sandoval, G. T., Jaimes, G. C., Barrios, M. C., Cespedes, C. & Velasco, H. M. SHOX gene and conserved noncofing element deletions/duplications in Colombian patients with idiopathic short stature. Mol. Genet. Genomic Med. 2, 95–102 (2014).
Fukami, M., Naiki, Y., Muroya, K., Hamajima, T., Soneda, S., Horikawa, R. et al. Rare pseudoautosomal copy-number variations involving SHOX and/or its flanking regions in individuals with and without short stature. J. Hum. Genet. 60, 553–556 (2015).
Tropeano, M., Howley, D., Gazzellone, M. J., Wilson, C. E., Ahn, J. W., Stavropoulos, D. J. et al. Microduplications at the pseudoautosomal SHOX locus in autism spectrum disorders and related neurodevelopmental conditions. J. Med. Genet. 53, 536–547 (2016).
Benito-Sanz, S., Gorbenko del Blanco, D., Huber, C., Thomas, N. S., Aza-Carmona, M., Bunyan, D. et al. Characterization of SHOX deletions in Léri-Weill dyschondrosteosis (LWD) reveals genetic heterogeneity and no recombination hotspots. Am. J. Hum. Genet. 79, 409–414 (2006b).
Potocki, L., Chen, K. S., Park, S. S., Osterholm, D. E., Withers, M. A., Kimonis, V. et al. Molecular mechanism for duplication 17p11.2 the homologous recombination reciprocal of the Smith-Magenis microdeletion. Nat. Genet. 24, 84–87 (2000).
Shaw, C. J., Bi, W. & Lupski, J. R. Genetic Proof of Unequal Meiotic Crossovers in Reciprocal Deletion and Duplication of 17p11.2. Am. J. Hum. Genet. 71, 1072–1081 (2002).
Bi, W., Park, S. S., Shaw, C. J., Withers, M. A., Patel, P. I. & Lupski, J. R. Reciprocal crossovers and a positional preference for strand exchange in recombination events resulting in deletion or duplication of chromosome 17p11.2. Am. J. Hum. Genet. 73, 1302–1315 (2003).
Acknowledgements
This work was supported in part by the following grants: Ministerio de Economia y Competitividad (MINECO) (SAF2003-05211, SAF2006-00663, SAF2009-08230, SAF2012-30871, SAF2015-66831-R), Ministerio de Sanidad (Fondo de Investigación Sanitaria, FIS PI06/90364, PI08/90270 and the Comunidad de Madrid ENDOSCREEN (S2010/BMD-2396). SB-S was awarded a postdoctoral CIBERER (Centro de Investigaciones en Red de Enfermedades Raras) fellowship and AB-M and MA-C a postdoctoral contract from Eli Lilly España (Proyecto Crecemos).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Journal of Human Genetics website
Rights and permissions
About this article
Cite this article
Benito-Sanz, S., Belinchon-Martínez, A., Aza-Carmona, M. et al. Identification of 15 novel partial SHOX deletions and 13 partial duplications, and a review of the literature reveals intron 3 to be a hotspot region. J Hum Genet 62, 229–234 (2017). https://doi.org/10.1038/jhg.2016.113
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2016.113
This article is cited by
-
Clinical, cytogenomic, and molecular characterization of isodicentric Y-chromosome and prediction of testicular sperm retrieval outcomes in azoospermic and severe oligozoospermic infertile men
Journal of Assisted Reproduction and Genetics (2022)
-
Unbalanced X;9 translocation in an infertile male with de novo duplication Xp22.31p22.33
Journal of Assisted Reproduction and Genetics (2019)
-
Identification of a limb enhancer that is removed by pathogenic deletions downstream of the SHOX gene
Scientific Reports (2018)
-
A novel missense mutation in the HECT domain of NEDD4L identified in a girl with periventricular nodular heterotopia, polymicrogyria and cleft palate
Journal of Human Genetics (2017)