Abstract
Human APOBEC3H (A3H) is a member of APOBEC cytidine deaminase family intensively constraining the HIV-1 replication. A3H is known to be polymorphic with different protein stability and anti-HIV-1 activity in vitro. We recently reported that A3H haplotypes composed of two functional polymorphisms, rs139292 (N15del) and rs139297 (G105R), were associated with the susceptibility to HIV-1 infection in Japanese. To confirm the association of A3H and HIV-1 infection in another ethnic group, a total of 241 HIV-1-infected Indian individuals and ethnic-matched 286 healthy controls were analyzed for the A3H polymorphisms. The frequency of 15del allele was high in the HIV-1-infected subjects as compared with the controls (0.477 vs 0.402, odds ratio (OR)=1.36, P=0.014). Haplotype analysis showed that the frequencies of 15del–105R was high (0.475 vs 0.400, OR=1.36, permutation P=0.037) in the HIV-1-infected subjects, confirming the association of A3H polymorphisms with the susceptibility to HIV-1 infection.
Similar content being viewed by others
Introduction
Human apolipoprotein B mRNA-editing catalytic polypeptide-3 (APOBEC3) gene family encodes cellular DNA cytosine deaminases that are involved in antiretroviral activity mediated through inhibition of viral replication.1 Expression of mRNA for a member of APOBEC3 family, APOBEC3H (A3H), is induced by interferon-α in target cells of HIV-1 infection,2, 3, 4 although it is difficult to detect cellular protein expression due to a low stability of A3H protein.5 The A3H protein is highly polymorphic with amino acid changes due to sequence variations including N15del (rs139292) and R18L in exon 2, R105G (rs139297) and K121E/D (rs139298) in exon 3, and E178D in exon 4, which are in linkage disequilibria with each other and composing of several different haplotypes.4, 5, 6, 7, 8 It has been demonstrated that A3H haplotypes I, III, IV and VI encode unstable A3H proteins, while haplotype II, V and VII, which share both 15N and 105R, produce stable protein and exert strong activity against HIV-1 replication in vitro.4, 5, 6, 7, 9, 10 Several studies in European descendant and African subjects have revealed that the susceptibility to HIV-1 infection and/or AIDS progression is controlled by human genetic factors.11, 12, 13 We have earlier reported that genetic factors such as polymorphisms in genes for HLA-B, TRIM5α and TIM1 are involved in the genetic regulation of susceptibility to HIV-1 infection or progression to AIDS in Asian populations including Japanese and Indian.14, 15, 16, 17, 18 More recently we reported that the A3H polymorphisms rs139292 and rs139297, and their haplotypes were associated with the susceptibility to HIV-1 infection in a Japanese population.19 In this study, we investigated the A3H polymorphisms in an Indian population to confirm the association in another ethnic group.
Materials and Methods
Blood DNA samples were obtained from 241 HIV-1-infected Indian patients and 286 healthy Indian volunteers in the related hospitals of the All India Institute of Medical Sciences, New Delhi, as described previously.17 Genotyping for A3H polymorphisms was performed by a fragment length analysis for rs139292 and RFLP analysis using FspI (New England Biolabs Japan, Tokyo, Japan) for rs139297, respectively, of PCR products amplified by using primer pairs described previously19 except that we used a VIC-labeled primer instead of 6-FAM-labeled primer in the fragment length analysis. Confirmation of genotyping data was done by direct sequencing of randomly selected (about 60%) samples. Strength of the association was expressed as odds ratio (OR) with 95% confidence interval (95% CI) calculated from two-by-two tables and statistical significance was evaluated by the χ2-test. Conformity of genotype distribution to Hardy–Weinberg equilibrium was tested by comparing the observed and expected genotype frequencies by using two-by-three tables and the χ2-test. Estimation of haplotype frequency and haplotype association study to obtain P-values by permutation test (n=1 000 000) were done by using the Haploview software (https://www.broadinstitute.org/scientific-community/science/programs/medical-and-population-genetics/haploview/haploview).20 Because this is a replication study of the association between the susceptibility to HIV-1 infection and recently reported HIV-1-infection-associated A3H polymorphisms, P-values were not corrected for multiple testing to avoid false-negative results (type II errors). P-values <0.05 were considered to be significant. Appropriate informed consent was given by each subject and the study protocol was approved by the ethics committees of the participating institutes: Medical Research Institute; Tokyo Medical and Dental University; and the All India Institute of Medical Sciences.
Results and Discussion
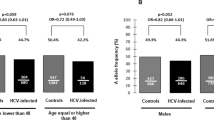
To investigate the association of A3H polymorphisms and susceptibility to HIV-1 infection in an Indian population, 241 patients and 286 controls were genotyped for rs139292 (N15del) and rs139297 (G105R). We found that minor allele frequencies for rs139292 (15del allele frequency) and rs139297 (105R allele frequency) were significantly higher in the patients than in the controls (patients vs controls: 0.477 vs 0.402; OR=1.36; 95% CI=1.06–1.73; P=0.014; and 0.519 vs 0.455; OR=1.29; 95% CI=1.01–1.65; P=0.038, respectively). The association with 15del allele and that with 105R allele was consistent with either of additive, dominant and recessive model, but was most likely fitting to the additive model, because ORs for homozygotes were nearly a square of OR for heterozygotes in both polymorphisms (Table 1).
Owing to the strong linkage disequilibrium between rs139292 and rs139297, A3H haplotypes could be inferred and categorized into three groups: hapA (defined by 15N and 105G); hapB (defined by 15N and 105R); and hapC (defined by 15del and 105R).19 We next conducted a haplotype association study by comparing estimated haplotype frequencies and permutation test by using the Haploview software. It was found that the frequency of 15del–105R haplotype (hapC) was significantly higher in the patients than in the controls (0.475 vs 0.400, OR=1.36, 95% CI=1.06–1.73, P=0.037). On the other hand, the frequency of 15N–105G haplotype (hapA) was low in the patients as compared with the controls although it was not statistically significant (0.479 vs 0.544, OR=0.77, 95% CI=0.61–0.98, P=0.081; Table 2).
The higher frequency of hapC in the patients is consistent with our previous finding in Japanese,19 thus confirming that the A3H hapC, of which protein expression is barely detectable in primary CD4+ T cells,10 is associated with the susceptibility to HIV-1 infection. Because hapB encodes a stable A3H protein,9 a slightly, though not significantly, decreased frequency of hapB found in Asian Indians might be explained by that the A3H of hapB haplotype exerted anti-HIV-1 activity in vivo. On the other hand, the frequency of hapA, which also encodes unstable A3H protein, was decreased in the Asian Indian patients (OR=0.77), as was found in the Japanese patients (OR=0.75).19 The reason for that the unstable A3H-encoding haplotype showed a possible resistance against HIV-1 infection remains to be solved in future studies.
In addition to the decreased ORs for haplotype A to similar extent in both ethnic groups, it is interesting to note that ORs for haplotype C were 1.36 and 1.33 in Indian and Japanese populations, respectively. Even though there was a considerable difference in the frequencies of hapA and hapC in the different ethnic groups, that is, 0.540 and 0.697 for hapA and 0.400 and 0.262 for hapC in the healthy Indian and Japanese controls, respectively, suggesting that the genetic controls for protection by hapA and susceptibility by hapC were similarly effective in both ethnic groups, even though there is a possibility that the variability in infected HIV-1 Vif sequences might be different in India and Japan, because the resistance to HIV-1 infection conferred by A3H is known to be dependent on the variations in Vif sequences.10
In conclusion, we report here that the A3H hapC (15del–105R) or 15del allele itself that are known to show relatively lower anti-HIV activity are associated with HIV-1 infection in Indians as was found in Japanese.
References
Desimmie, B. A., Delviks-Frankenberrry, K. A., Burdick, R. C., Qi, D., Izumi, T. & Pathak, V. K. Multiple APOBEC3 restriction factors for HIV-1 and one Vif to rule them all. J. Mol. Biol. 426, 1220–1245 (2014).
OhAinle, M., Kerns, J. A., Malik, H. S. & Emerman, M. Adaptive evolution and antiviral activity of the conserved mammalian cytidine deaminase APOBEC3H. J. Virol. 80, 3853–3862 (2006).
Harari, A., Ooms, M., Mulder, L. C. & Simon, V. Polymorphisms and splice variants influence the antiretroviral activity of human APOBEC3H. J. Virol. 83, 295–303 (2009).
Tan, L., Sarkis, P. T., Wang, T., Tian, C. & Yu, X. F. Sole copy of Z2-type human cytidine deaminase APOBEC3H has inhibitory activity against retrotransposons and HIV-1. FASEB J. 23, 279–287 (2009).
OhAinle, M., Kerns, J. A., Li, M. M., Malik, H. S. & Emerman, M. Antiretroelement activity of APOBEC3H was lost twice in recent human evolution. Cell Host Microbe 4, 249–259 (2008).
Wang, X., Abudu, A., Son, S., Dang, Y., Venta, P. J. & Zheng, Y. H. Analysis of human APOBEC3H haplotypes and anti-human immunodeficiency virus type 1 activity. J. Virol. 85, 3142–3152 (2011).
Ooms, M., Majdak, S., Seibert, C. W., Harari, A. & Simon, V. The localization of APOBEC3H variants in HIV-1 virions determines their antiviral activity. J. Virol. 84, 7961–7969 (2010).
Zhen, A., Du, J., Zhou, X., Xiong, Y. & Yu, X. F. Reduced APOBEC3H variant anti-viral activities are associated with altered RNA binding activities. PLoS ONE 7, e38771 (2012).
Zhen, A., Wang, T., Zhao, K., Xiong, Y. & Yu, X. F. A single amino acid difference in human APOBEC3H variants determines HIV-1 Vif sensitivity. J. Virol. 84, 1902–1911 (2010).
Refsland, E. W., Hultquist, J. F., Luengas, E. M., Ikeda, T., Shaban, N. M., Law, E. K. et al. Natural polymorphisms in human APOBEC3H and HIV-1 Vif combine in primary T lymphocytes to affect viral G-to-A mutation levels and infectivity. PLoS Genet. 11, e1004761 (2014).
LeClerc, S., Limou, S., Coulonges, C., Carpentier, W., Dina, C., Taing, L. et al. Genomewide association study of a rapid progression cohort identifies new susceptibility alleles for AIDS (ANRS Genomewide Association Study 03). J. Infect. Dis. 200, 1194–1201 (2009).
Chinn, L. W., Tang, M., Kessing, B. D., Lautenberger, J. A., Troyer, J. L., Malasky, M. J. et al. Genetic associations of variants in genes encoding HIV-dependency factors required for HIV-1 infection. J. Infect. Dis. 202, 1836–1845 (2010).
LeClerc, S., Coulonges, C., Delaneau, O., VanManen, D., Herbeck, J. T., Limou, S. et al. Screening low-frequency SNPS from genome-wide association study reveals a new risk allele for progression to AIDS. J. Acquir. Immune Defic. Syndr. 56, 279–284 (2011).
Munkanta, M., Terunuma, H., Takahashi, M., Hanabusa, H., Miura, T., Ikeda, S. et al. HLA-B polymorphism in Japanese HIV-1-infected long-term surviving hemophiliacs. Viral Immunol. 18, 500–505 (2005).
Nakajima, T., Nakayama, E. E., Kaur, G., Terunuma, H., Mimaya, J. I., Ohtani, H. et al. Impact of novel TRIM5alpha variants, Gly110Arg and G176del, on the anti-HIV-1 activity and the susceptibility to HIV-1 infection. AIDS 23, 2091–2100 (2009).
Wichukchinda, N., Nakajima, T., Saipradit, N., Nakayama, E. E., Ohtani, H., Rojanawiwat, A. et al. TIM1 haplotype may control the disease progression to AIDS in a HIV-1-infected female cohort in Thailand. AIDS 24, 1625–1631 (2010).
Sharma, G., Ohtani, H., Kaur, G., Naruse, T. K., Sharma, S. K., Vajpayee, M. A. et al. Status of TIM-1 exon 4 haplotypes and CD4+T cell counts in HIV-1 seroprevalent North Indians. Hum. Immunol. 74, 163–165 (2013).
Nakayama, E. E., Nakajima, T., Kaur, G., Mimaya, J. I., Terunuma, H., Mehra, N. et al. A naturally occurring single amino acid substitution in human TRIM5α linker region affects its anti-HIV type 1 activity and susceptibility to HIV type 1 infection. AIDS Res. Hum. Retroviruses 29, 919–924 (2013).
Sakurai, D., Iwatani, Y., Ohtani, H., Naruse, T. K., Terunuma, H., Sugiura, W. et al. APOBEC3H polymorphisms associated with the susceptibility to HIV-1 infection and AIDS progression in Japanese. Imunogenetics 67, 253–257 (2015).
Barrett, J. C., Fry, B., Maller, J. & Daly, M. J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21, 263–265 (2005).
Acknowledgements
We thank Ms N Ohkubo and Y Ueda for their technical assistance. This work was supported in part by research grants from the Ministry of Health, Labor and Welfare, Japan; Grant-in-Aids for Scientific research from the Ministry of Education, Culture, Sports, Science, and Technology, Japan; a grant from the Indian Council of Medical Research; and by grants for India–Japan Cooperative Science Program from Japan Society for the Promotion of Science (JSPS), Japan and Department of Science and Technology (DST). This work was also supported by a support for women researchers from the Tokyo Medical and Dental University and the Joint Usage/Research Program of Medical Research Institute, Tokyo Medical and Dental University.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Naruse, T., Sakurai, D., Ohtani, H. et al. APOBEC3H polymorphisms and susceptibility to HIV-1 infection in an Indian population. J Hum Genet 61, 263–265 (2016). https://doi.org/10.1038/jhg.2015.136
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2015.136