Abstract
With homozygosity mapping we have identified two large homozygous regions on chromosome 3q13.11–q13.31 and chromosome 19p13.3–q31.32 in a large Pakistani family suffering from autosomal recessive nonsyndromic hearing impairment (arNSHI). The region on chromosome 19 overlaps with the previously described deafness loci DFNB15, DFNB72 and DFNB95. Mutations in GIPC3 have been shown to underlie the nonsyndromic hearing impairment linked to these loci. Sequence analysis of all exons and exon–intron boundaries of GIPC3 revealed a homozygous canonical splice site mutation, c.226-1G>T, in GIPC3. This is the first mutation described in GIPC3 that affects splicing. The c.226-1G>T mutation is located in the acceptor splice site of intron 1 and is predicted to affect the normal splicing of exon 2. With a minigene assay it was shown to result in the use of an alternative acceptor site in exon 2, resulting in a frameshift and a premature stop codon. This study expands the mutational spectrum of GIPC3 in arNSHI.
Similar content being viewed by others
Main
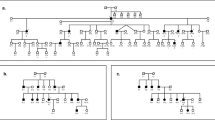
Hearing impairment is the most common sensory disorder worldwide. Pakistan has a high rate of consanguineous marriages and therefore this population is very suitable for collecting large families for genetic studies in recessive disorders. Many genes and loci for arNSHI have been mapped in families of Pakistani origin (hereditary hearing loss homepage). It has been shown that mutations in GJB2 (MIM *121011) are the most prevalent cause of deafness in the Pakistani population; mutations in this gene cause deafness in about 45–50% of the families.1 A large consanguineous Pakistani family (16DFN, Figure 1a) with prelingual arNSHI, residing in Southern Punjab, was selected for genetic screening. The study was approved by the human subjects review board of the Institute of Biomedical and Genetic Engineering, Islamabad, Pakistan. Peripheral blood was sampled from nine members of the three-generation-family, and DNA was extracted using a phenol chloroform method.2 Mutations in the previously reported most frequently implicated genes in arNSHI, GJB2 and MYO15A (MIM *602666) were excluded;1 GJB2 was analyzed via Sanger sequencing of both exons and MYO15A by genotyping microsatellite markers D17S1824 and D17S2196. Subsequently, the samples of the affected individuals IV.2, IV.4 and IV.5 were genotyped using the Human Omniexpress array (Illumina, San Diego, CA, USA) according to the manufacturer’s protocol. Homozygous regions were determined using the freely available Homozygosity Mapper software (www.homozygositymapper.org).3 In total, three homozygous regions larger than 1 Mb were shared by the three affected individuals (Figure 1b). A small homozygous region of 1.08 Mb was identified on chromosome 6p22.1 flanked by rs2531817 and rs362529 (chr6: 28,448,894-29,530,868, hg19). Further, two large homozygous regions were identified on chromosome 3q13.11–q13.31 and chromosome 19p13.3-q13.32. The region on chromosome 3, flanked by rs1994818 and rs1347060, encompassed 13.4 Mb (chr3:103,026,304-116,455,442, hg19). The region on chromosome 19 encompassed 45.3 Mb (chr19:3,414,088-49,092,430, hg19) flanked by rs12975319 and rs12984929. The homozygous region on chromosome 3 did not contain or overlap with a known deafness locus or gene. However, several loci for arNSHI have been mapped to chromosome 19. The homozygous region identified in the present family was much larger than any of the loci described so far on chromosome 19. The present region contains the previously described DFNB68 and DFNB81 loci and partially overlaps with the DFNB15, DFNB72 and DFNB95 loci.4, 5, 6, 7, 8 For DFNB68 and DFNB81 no disease-associated mutations have been reported so far. For DFNB15, DFNB72 and DFNB95 mutations in GIPC3 (MIM *608792) were found to be causative for the disease.5,7 As GIPC3 resides within the homozygous region of chromosome 19, all exons and exon–intron boundaries were screened by Sanger sequencing in affected individual IV.2. Primer sequences and PCR conditions have been described previously.5 The previously reported mutations (Supplementary Table 2) were not present in affected individual IV.2. However, a canonical splice site mutation was identified in intron 1 at genomic position Chr19(GRCh37):g.3586492G>T (c. 226-1G>T, reference sequence NM_133261.2) segregating with the hearing impairment in the family (Figure 1a and Supplementary Figure 1). The altered nucleotide is highly conserved and this variation has not been reported in any database so far, not even in the Exome Variant Server, which demonstrates the uniqueness of this variant.
The c.226-1G>T mutation abolishes the original splice acceptor site of intron 1, going from a score of 76.0 (calculated by the Human Splicing Finder, range 1–100) in the wild-type situation to no score at all for the mutation (Supplementary Figure 2). This will affect the normal splicing of exon 2; however, the resulting effect of the mutation is difficult to predict. One possibility is the use of an alternative splice acceptor site located within exon 2. This alternative splice acceptor site has a consensus value of 83.4, which is even higher than that of the original splice acceptor site (76.0, Supplementary Figure 2). Another possibility is that exon 2 will be skipped completely. This exon consists of 186 nucleotides and encodes 62 amino acids. To determine the effect of the c.226-1G>T mutation on splicing, a minigene approach was used as previously described.1 Primer sequences to amplify a fragment containing exon 2 and 3 of GIPC3 are shown in Supplementary Table 1. The minigene assay revealed correct splicing of the wild-type GIPC3 exon 2, whereas the c.226-1G>T mutation resulted in the use of the predicted alternative acceptor site in exon 2 (Figure 2). This leads to absence of the first 23 nucleotides of exon 2 in the mRNA and causes a frameshift and a premature stop codon. The predicted truncated protein does not have the PDZ domain and thus loses its function.
Splicing effect of the c.226-1G>T mutation (a) An agarose gel containing Reverse transcription-PCR products detected from HEK293T cells transfected with the wild-type and mutant minigene construct. (b) A schematic representation of the identified splicing products. The RT-PCR products were verified with sequence analysis. (c) Electropherogram of the partial cDNA sequence of RNA derived from cells transfected with the pCI-NEO with either the mutant or wild-type GIPC3 exon 2. The c.226-1G>T mutation leads to the use of an alternative splice acceptor site in exon 2 and to the deletion of the first 23 nucleotides of exon 2.
In total 12 mutations have been described in GIPC3, including the mutation identified in the current study, which all cause arNSHI (Supplementary Table 2).5,7,9, 10, 11 Most of these mutations were found in the Pakistani population thus showing the major contribution of this gene to deafness in this population. The mutations described for GIPC3 include three truncating mutations, and nine missense mutations. Two missense mutations have been reported so far in the PDZ domain of GIPC3.7,11 The other disease-related mutations described so far are in regions of unknown function or in low complexity regions (Supplementary Table 2).5,7,10
Different degrees of hearing impairment have been reported in families with GIPC3 mutations, ranging from mild-to-severe to profound.5,7,9, 10, 11 No clear relation is evident between the type or location of the mutations in GIPC3 and the degree of hearing loss. Pure-tone air conduction audiometry was available from affected individuals IV.2 and IV.4 of family 16DF. Both individuals show severe to profound hearing impairment affecting all frequencies at the age of 17 and 14, respectively (Figure 3). The hearing impairment was self-reported to be congenital and non-progressive. Progression of hearing loss has been reported in one individual of a Dutch family, who had a mutation in GIPC3.5
Mouse studies have suggested that GIPC3 is required for the postnatal maturation of hair bundles in the inner ear and long-term survival of hair cells and spiral ganglion.5 Gipc3 mutant mice showed defects in mechanotransduction in both inner and outer hair cells that were associated with defects in potassium channel activity.5 Identification of novel mutations in GIPC3 may further elucidate the role of GIPC3 in various pathways of sound transduction and processing. Also, it may aid to further define the phenotype associated with GIPC3 mutations.
References
Shafique S, Siddiqi S, Schraders M, Oostrik J, Ayub H, Bilal A et al. Genetic spectrum of autosomal recessive non-syndromic hearing loss in pakistani families. PLoS ONE 2014; 9: e100146.
Sambrook J, Fritsch EF & Maniatis T Analysis and Cloning of Eukaryotic Genomic DNA. Molecular Cloning: A Laboratory Manual. (Cold Spring Harbour Laboratory Press, New York, 1989).
Seelow D, Schuelke M, Hildebrandt F & Nurnberg P HomozygosityMapper—an interactive approach to homozygosity mapping. Nucleic Acids Res. 2009; 37: W593–W599.
Ain Q, Nazli S, Riazuddin S, Jaleel AU, Riazuddin SA, Zafar AU et al. The autosomal recessive non-syndromic deafness locus DFNB72 is located on chromosome 19p13.3. Hum. Genet. 2007; 122: 445–450.
Charizopoulou N, Lelli A, Schraders M, Ray K, Hildebrand MS, Ramesh A et al. Gipc3 mutations associated with audiogenic seizures and sensorineural hearing loss in mouse and human. Nat. Commun. 2011; 2: 201.
Chen A, Wayne S, Bell A, Ramesh A, Srisailapathy CR, Scott DA et al. New gene for autosomal recessive non-syndromic hearing loss maps to either chromosome 3q or 19p. Am. J. Med. Genet. 1997; 71: 467–471.
Rehman AU, Gul K, Morell RJ, Lee K, Ahmed ZM, Riazuddin S et al. Mutations of GIPC3 cause nonsyndromic hearing loss DFNB72 but not DFNB81 that also maps to chromosome 19p. Hum. Genet. 2011; 130: 759–765.
Santos RL, Hassan MJ, Sikandar S, Lee K, Ali G, Martin PE Jr. et al. DFNB68, a novel autosomal recessive non-syndromic hearing impairment locus at chromosomal region 19p13.2. Hum. Genet. 2006; 120: 85–92.
Diaz-Horta O, Duman D, Foster J 2nd, Sirmaci A, Gonzalez M, Mahdieh N et al. Whole-exome sequencing efficiently detects rare mutations in autosomal recessive nonsyndromic hearing loss. PLoS ONE 2012; 7: e50628.
Ramzan K, Al-Owain M, Allam R, Berhan A, Abuharb G, Taibah K et al. Homozygosity mapping identifies a novel GIPC3 mutation causing congenital nonsyndromic hearing loss in a Saudi family. Gene 2013; 521: 195–199.
Sirmaci A, Edwards YJ, Akay H & Tekin M Challenges in whole exome sequencing: an example from hereditary deafness. PLoS ONE 2012; 7: e32000.
Acknowledgements
We are thankful to the Higher Education Commission, Pakistan for providing Post Doc scholarship to SS to conduct this study in the group of Prof. H. Kremer in the Netherlands. This work was financially supported by a grant from ZorgOnderzoek Nederland / Medische Wetenschappen (40-00812-98-09047, to H.K.).
Web resources. The URLs for the data presented herein are as follows: The Hereditary Hearing loss Homepage, http://hereditaryhearingloss.org/. HomozygosityMapper, http://www.homozygositymapper.org/. Polymorphism Phenotyping (Polyphen-2), http://genetics.bwh.harvard.edu/pph2/. Sorting Intolerant from Tolerant (SIFT), http://sift.jcvi.org/. UCSC Human Genome Database, Build hg19, http://www.genome.ucsc.edu/. Exome variant Server, http://evs.gs.washington.edu/EVS/. 1000 Genome projects, http://browser.1000genomes.org/. National Center for Biotechnology Information (NCBI), http://www.ncbi.nlm.nih.gov/
Author contributions: SS and JO performed the experiments, SM and MI provided the family for the study. SS did the data analysis. HK provided all reagents/software. SS wrote the paper. RQ, AM, HK and MS supervised the work and critically read the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Siddiqi, S., Ismail, M., Oostrik, J. et al. A canonical splice site mutation in GIPC3 causes sensorineural hearing loss in a large Pakistani family. J Hum Genet 59, 683–686 (2014). https://doi.org/10.1038/jhg.2014.86
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2014.86
This article is cited by
-
Low incidence of GIPC3 variants among the prelingual hearing impaired from southern India
Journal of Genetics (2020)