Abstract
Recent reviews discussed the critical roles of apoptosis in human spermatogenesis and infertility. These reviews highlight the FasL-induced caspase cascade in apoptosis lending importance to our discovery of the pseudogene status of the Lfg5 gene in modern humans, Neanderthal and the Denisovan. This gene is a member of the ancient and highly conserved apoptosis Lifeguard family. This pseudogenization is the result of a premature stop codon at the 3′-end of exon 8 not found in any other ortholog. With the current exception of the domesticated bovine and buffalo, Lfg5′s expression in mammals is testis-specific. A full analysis of this gene, its phylogenetic context and its recent hominin changes suggest its inactivation was likely under selection in human evolution.
Similar content being viewed by others
Main
Human Lfg5 is currently identified as Hs.641506.1, 2, 3, 4, 5 In fact, it is an ortholog of mouse Tmbim7/4930511M11Rik and cow Tmbim1b.6 (Supplementary Figure S1, Supplementary materials. Its predicted mRNA and protein sequences are shown in Supplementary Figure S2 and Supplementary materials) To put the human Lfg5 pseudogene in context, an extensive phylogenetic analysis of extant Lifeguard, LFG, genes among the opisthokonts, chromalveolates, stramenopiles, excavates and plant clades was carried out (Figure 1; Supplementary Figure S1, Supplementary materials). The most probable ancestral family progenitor, appears to have been a Lfg4-like progenitor of Lfg4 and Lfg1/5 subgroups and precedes the divergence of those clades. Animals and alveolates contain both Lfg4- and Lfg1/5-like genes; green plants, red and brown algae, stramenopiles, fungi, choanoflagellates and sponges, have Lfg4 or its clear derivatives; lineages of green algae genes remain uncertain. The family expanded by a series of duplications and subsequent modifications (Figure 1; Supplementary Figure S1, Supplementary materials). The Lfg1/Lfg5 progenitor produced the earliest forms of Lfg1 and Lfg5. Lfg1 underwent a double duplication in vertebrates generating the precursors of Lfg2 and Lfg3.
Reconstructed phylogeny of the Lifeguard gene family. This tree represents a summary of a full analysis of over eight hundred available and identifiable LFG family homologs. For the full LFG family eukaryotic tree analysis, see Supplementary materials and Supplementary Figure S1. Current family members are indicated in black next to each clade. Within Eumetazoa, presumed gene losses are in red on their respective branches. Duplications observed only in certain species within a lineage are indicated in blue, and analogous gene losses are in gray (for example, some Nematoda duplicated Lfg1, whereas most Sauropsida lost Lfg5). The Homo-specific Lfg5 pseudogenization is highlighted in the purple circle. Phylogenetic reconstructions using the full available set of the LFG family members did not resolve the most probable ancestral precursor as either that of an Lfg4 or Lfg1/5 precursor. See Supplementary Materials for the full phylogenetic analysis and the methods used.
Within all eutherian mammalian genomes, five paralogs can be identified, ergo Lfg4, Lfg1, Lfg2, Lfg3 and Lfg5. There are distinctive sequence patterns for each of these five subfamilies among the extant animals,6 the most characteristic amino-acid pattern is found near the C-terminal: ‘[SN]P[ED][ED]YX(9)D’. There is high level of conservation within these subfamilies, in their amino-acid sequences, their gene structure (Figure 2) and their genomic context, for example, Lfg5 is found between the genes, ankib1 and gatad1, in a common syntenic block from reptiles through eutherian mammals.
A number of alternate names have been assigned to the LFG family genes, making the literature potentially confusing.6 The LFG genes have often been assumed homologs of other small six- or seven-transmembrane scaffolded proteins, such as the Bcl/Bax inhibitor family by virtue of their similar transmembrane structural and shared apoptotic roles.7 However, the LFG consensus lacks the conserved BI1 C-terminal and respective functions as shown for S. cerevisiae Ynl305c/Lfg4.8 Although some very distant relationship or even convergence cannot be ruled out, the LFGs always phylogenetically cluster independent of Bax-motif containing genes, as far back as the root of all animals, if not all extant eukaryotes, qualifying them as an independent eukaryotic gene family.
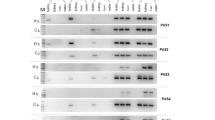
A review of literature indicates apoptosis and its regulation as a common functional feature among all of the animal Lifeguard proteins.6, 9, 10, 11, 12, 13 Originally, Lfg5 was identified bioinformatically in bovine, mouse and human.6 The Lfg5 expression in mammals is postnatal and testis-restricted in all but the domesticated cow and buffalo where it is not sex dimorphic. The expression of two of the five Lfg5 paralogs in fruit fly is testis-restricted in adults6 (UniGene and FlyBase 2012). Lfg5 EST expression in the Anolis lizard is also testis-restricted. Lfg5 in mouse is expressed in leptotene–zygotene spermatocytes,5 coinciding with spermatocyte crossing blood–testis barrier into the adluminal compartment of seminiferous tubules, peaking during 18–26 days postpartum.4 This expression is at 22% of Stk31, the highest expressed mouse testis-specific gene.14 Lfg5 is expressed in the bonobo again in a testis-specific fashion.15, 16 In humans, Neanderthal and Denisovan Lfg5 orthologs, a conserved premature stop codon is found at the 3′-end of exon 8, placing it in the fifth loop of the protein (Supplementary Figure S3, Supplementary materials). In addition, another premature stop codon at the 2nd triplet upstream of the conserved one is found in the Neanderthal Lfg5 only. Full-length human Lfg5 mRNA is not observed in any human EST data currently available (NCBI-GEO database) in contrast with the high expression of the complete Lfg5 transcript in bonobo (marmoset and chimpanzee), mouse and dog testis (Figure 3). There is very-low-level detectable Lfg5 mRNA expression in human but it is only fragmentary or misspliced. A verification of the dominant misspliced transcript was obtained by PCR sequencing of human testis complementary DNA (Supplementary Figure S2, Supplementary materials).
The Lfg5 pseudogene status in humans, Neanderthal and Denisovan line of descent is curious, given the gene’s high sequence, structure, synteny and testis expression conservation in mammals. This shortened and apparent inactivated status of Lfg5, hence predates the H. sapiens-Neanderthal divergence, some 300 000 years ago.17 This must be considered a minimum age, as there is no support for an interbreeding common contig in this region on chromosome seven.18 Could this hominid pseudogenization of a mammalian testis-specific apoptotic gene even date as far back as H. heidelbergensis or H. erectus?
Lfg5 has been lost in extant birds and some reptiles, suggesting Lfg5’s apoptotic spermatocyte function is replaceable. Studies in humans have discussed the involvement of Bcl-x and Bax in testis spermatogenesis.19, 20 These same genes are also active in mouse spermatogenesis perhaps complementing Lfg5, but not replacing it. Perhaps Lfg5’s human function has not been replaced. The alignment of the Lfg5 DNA exons among the primates (Supplementary Figure S3, Supplementary materials) provides an analysis of the ratio of synonymous to nonsynonymous changes (Figure 4). The dn/ds Nei-Gojobori statistic after the Orangutan divergence is significantly greater than unity supporting the idea of positive selection. Given Lfg5 is fully and uniquely expressed in the testis of our close relatives, the chimpanzee and bonobo, what kind of selection might there have been?
The sequential distribution of nonsynonymous (first) versus synonymous (second). mutations are displayed along each line of descent to human. The bracketed numbers displayed under the lines are the Nei-Gojobori statistic. The identified fixed substation counts have been placed on the accepted primate tree topology,17 that is identical to that produced using the protein sequences of Lfg5 and/or Lfg4, accept for relative branch lengths. The pig, Sus scrofa domesticus, was used as a mammalian outgroup reference. These data were derived from the full phylogenetic analysis of all clearly identified Lfg5s in all animals by full multi-alignment and Phyml tree analysis, using a null model (fixed KaKs along the whole tree). The statistics were then computed comparing the consensus sequences at the relevant ancestral nodes. The full tree and detailed analysis is available in the Supplementary materials.
Given the recent studies showing the low percentage of fully functional human sperm21 and its relationship to apoptosis,3 there is a purely hypothetical possibility that reduced fertility was under some form of selection. Clearly in humans there is a reduction, if not loss, of most outward signs of female’s ovulation timing, potentially allowing more continuous sexual interaction without changing the female fertility. Is there any connection on the male’s side allowing reduce fertility? It is clear that there is significant size and development timing differences in the testis between the chimpanzee, gorilla and extant humans that is correlated with the degree of female promiscuity and the frequency of her fertile period.22, 23 Whether any of this is related to the level of active human sperm or the loss of Lfg5, is currently at the level of speculation, suggesting opportunities for more research.
References
Tripathi, R, Mishra, DP & Shaha, C Male germ cell development: turning on the apoptotic pathways. Reprod. Immunol. 83, 31–35 (2009).
Shaha, C, Tripathi, R & Mishra, DP Male germ cell apoptosis: regulation and biology. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 365, 1501–1515 (2010).
Shukla, KK, Mahdi, AA & Rajender, S Apoptosis, spermatogenesis and male infertility. Front. Biosci. (Elite Ed) 4, 746–754 (2012).
Schultz, N, Hamra, FK & Garbers, DL A multitude of genes expressed solely in meiotic or postmeiotic spermatogenic cells offers a myriad of contraceptive targets. Proc. Natl Acad. Sci. USA 100, 12201–12206 (2003).
Kogo, H, Kowa-Sugiyama, H, Yamada, K, Bolor, H, Tsutsumi, M, Ohye, T et al. Screening of genes involved in chromosome segregation during meiosis I: toward the identification of genes responsible for infertility in humans. J. Hum. Genet. 55, 293–299 (2010).
Hu, L, Smith, TF & Goldberger, G LFG: a candidate apoptosis regulatory gene family. Apoptosis 14, 1255–1265 (2009).
Carrara, G, Saraiva, N, Johnson, BF, Gubser, C & Smith, GL A six transmembrane topology for Golgi anti-apoptotic protein (GAAP) and Bax Inhibitor 1 (BI-1) provides a model for the transmembrane Bax inhibitor containing motif (TMBIM) family. J. Biol. Chem. 287, 15896–15905 (2012).
Lisbona, F, Rojas-Rivera, D, Thielen, P, Zamorano, S, Todd, D, Martinon, F et al. BAX inhibitor-1 is a negative regulator of the ER stress sensor IRE1alpha. Mol. Cell 33, 679–691 (2009).
Henke, N, Lisak, DA, Schneider, L, Habicht, J, Pergande, M & Methner, A The ancient cell death suppressor BAX inhibitor-1. Cell Calcium 50, 251–260 (2011).
Kim, JH, Lee, ER, Jeon, K, Choi, HY, Lim, H, Kim, SJ et al. Role of BI-1 (TEGT)-mediated ERK1/2 activation in mitochondria-mediated apoptosis and splenomegaly in BI-1 transgenic mice. Biochim. Biophys. Acta 1823, 876–888 (2012).
Saraiva, N, Prole, DL, Carrara, G, Maluquer de Motes, C, Johnson, BF, Byrne, B et al. Human and viral Golgi anti-apoptotic proteins (GAAPs) oligomerize via different mechanisms and monomeric GAAP inhibits apoptosis and modulates calcium. J. Biol. Chem. 288, 13057–13067 (2012).
Weis, C, Hückelhoven, R & Eichmann, R LIFEGUARD proteins support plant colonization by biotrophic powdery mildew fungi. J. Exp. Bot. 64, 3855–3867 (2013).
Rojas-Rivera, D, Armisén, R, Colombo, A, Martínez, G, Eguiguren, AL, Díaz, A et al. TMBIM3/GRINA is a novel unfolded protein response (UPR) target gene that controls apoptosis through the modulation of ER calcium homeostasis. Cell Death. Differ. 19, 1013–1026 (2012).
Wang, PJ, McCarrey, JR, Yang, F & Page, DC An abundance of X-linked genes expressed in spermatogonia. Nat. Genet. 27, 422–426 (2001).
Brawand, D, Soumillon, M, Necsulea, A, Julien, P, Csárdi, G, Harrigan, P et al. The evolution of gene expression levels in mammalian organs. Nature 478, 343–348 (2011).
Prado-Martinez, J, Sudmant, PH, Kidd, JM, Li, H, Kelley, JL, Lorente-Galdos, B et al. Great ape genetic diversity and population history. Nature 499, 471–475 (2013).
Langergraber, KE, Prüfer, K, Rowney, C, Boesch, C, Crockford, C, Fawcett, K et al. Generation times in wild chimpanzees and gorillas suggest earlier divergence times in great ape and human evolution. Proc. Natl Acad. Sci. USA 109, 15716–15721 (2012).
Eriksson, A & Manica, A Effect of ancient population structure on the degree of polymorphism shared between modern human populations and ancient hominins. Proc. Natl Acad. Sci. USA 109, 13956–13960 (2012).
Oldereid, NB, Angelis, PD, Wiger, R & Clausen, OP Expression of Bcl-2 family proteins and spontaneous apoptosis in normal human testis. Mol. Hum. Reprod. 7, 403–408 (2001).
Ketola, I, Toppari, J, Vaskivuo, T, Herva, R, Tapanainen, JS & Heikinheimo, M Transcription factor GATA-6, cell proliferation, apoptosis, and apoptosis-related proteins Bcl-2 and Bax in human fetal testis. J. Clin. Endocrinol. Metab. 88, 1858–1865 (2003).
Sousa, AP, Amaral, A, Baptista, M, Tavares, R, Caballero Campo, P, Caballero Peregrín, P et al. Not all sperm are equal: functional mitochondria characterize a subpopulation of human sperm with better fertilization potential. PLoS One 6, e18112 (2011).
Gray, PB & Garcia, JR Evolution and Human Sexual Behavior (Harvard University Press, Cambridge, MA, USA, 2013).
Gray, PB Evolution and human sexuality. Am. J. Phys. Anthropol. 152 (Suppl 57), 94–118 (2013).
Acknowledgements
MM had support from the CRG in Barcelona. GG dedicates his efforts to the memory of his parents Rabbi Shimshon and Elka Goldberger.
Author information
Authors and Affiliations
Additional information
Supplementary Information accompanies the paper on Journal of Human Genetics website
Rights and permissions
About this article
Cite this article
Mariotti, M., Smith, T., Sudmant, P. et al. Pseudogenization of testis-specific Lfg5 predates human/Neanderthal divergence. J Hum Genet 59, 288–291 (2014). https://doi.org/10.1038/jhg.2014.6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2014.6