Abstract
Chronic obstructive pulmonary disease (COPD) is a complex, multifactorial disease. Although smoking is a main risk factor for obstructive impairment, not all smokers develop this critical disease. We conducted a genome-wide association study to identify the association between genetic variants and pulmonary function and also examined how these variants relate to lung impairment in accordance with smoking behaviors. Using two community-based cohorts, the Ansan cohort (n=4319) and the Ansung cohort (n=3674), in the Korean Genome Epidemiology Study, we analyzed the association between genetic variants (single-nucleotide polymorphisms and haplotypes) and the ratio of FEV1 to FVC (FEV1/FVC) using multivariate linear regression models. Similar analyses were conducted after stratification by smoking status. Four genome-wide significant signals in the FAM13A gene (the strongest signal at rs2609264, P=1.76 × 10−7 in a combined set) were associated with FEV1/FVC. For the association with ratio, the effect size in the CTGA haplotype (risk haplotype) was −0.57% (s.e., 0.11; P=2.10 × 10−7) as compared with the TCAG haplotype (reference haplotype) in a combined set. There was also a significant interaction of FAM13A haplotypes with heavy smoking on FEV1/FVC (P for interaction=0.028). We confirmed the previously reported association of FAM13A in 4q22.1 with pulmonary function. The FAM13A haplotypes also interacted with heavy smoking to affect the risk of reduced pulmonary function.
Similar content being viewed by others
Introduction
A pulmonary function test provides an accurate and objective assessment of lung impairment and can obtain adequate information for the diagnosis of chronic obstructive pulmonary disease (COPD).1 COPD, a disease characterized by obstructive ventilatory impairment, is one of the most common respiratory diseases and one of the leading causes of mortality worldwide.2, 3, 4 Forced expiratory volume in the first second (FEV1), forced vital capacity (FVC) and the ratio of FEV1 to FVC (FEV1/FVC) are the major spirometric parameters.1 An obstructive ventilatory defect presents a disruption in the balance between maximal airflow of the lung and its capacity. It means airway narrowing during exhalation and is defined by a reduced FEV1/FVC.5
COPD is a complex disease that is influenced by multifactorial environmental traits, such as old age, infection, presence of asthma and cigarette smoking.6, 7 Although smoking is the main risk factor for COPD, not all smokers develop this critical disease. Thus, it has been suggested that genetic factors may explain why some individuals are more susceptible than others. Several family studies have affirmed genetic influences on respiratory impairment, but little is known about which particular gene is involved.8, 9, 10
Recently, multiple loci associated with pulmonary function and/or risk of COPD were identified using genome-wide association (GWA) analyses.11, 12, 13, 14, 15, 16, 17, 18, 19 GWA studies (GWASs) for meta-analyses among four populations of the Cohorts for Heart and Aging Research in Genomic Epidemiology consortium reported that FEV1/FVC is associated with chromosome 4. In particular, the chromosome 4q31 region near HHIP and the 4q22.1 region in FAM13A were related to FEV1/FVC.11, 14, 15, 16, 17, 19 Some case–control studies replicated this finding and furthermore revealed that variants in FAM13A are associated with a higher risk of COPD, especially in smokers.16, 20, 21 However, the biological mechanism explaining how the interaction of single-nucleotide polymorphisms (SNPs) and environmental factors (for example, smoking status) increases the prevalence of reduced pulmonary function and/or COPD is still unclear.
In this study, we performed GWAS on the pulmonary function of two Korean population-based cohorts as part of the Korean Genome Epidemiology Study (KoGES). We investigated which genetic factors might contribute to pulmonary function as measured by FEV1/FVC and examined how they related to lung impairment in accordance with smoking behaviors.
Materials and methods
Study design and population cohorts
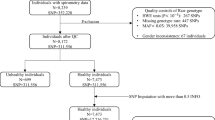
This study analyzed GWAS data from two independent cohorts from Ansan and Ansung that are included in the KoGES. Both cohorts enrolled Korean men and women aged 39–70 years in 2001–2002 to conduct a prospective investigation. Detailed information on participant recruitment is available elsewhere.22, 23, 24 A total of 5020 participants from Ansan and 5018 participants from Ansung took part in the baseline study from 2001 to 2003. Cohort members underwent a comprehensive health examination and a questionnaire-based interview, and biospecimens for assays were collected by health professionals at each study site. A questionnaire included demographic characteristics, lifestyle choices and medical history. Pulmonary function tests were performed by a skilled technician using a portable spirometer (Vmax-2130, Sensor Medics, Yorba Linda, CA, USA) according to standardized protocols of the American Thoracic Society.25 All participants performed pre-bronchodilator spirometry test until completing at least three repeated measurements and an acceptable measure was determined when the differences between the largest and the next largest FVC and FEV1 values were within 0.15 l. Calibration and quality control of spirometric examinations were also performed regularly based on American Thoracic Society guidelines.25
The Korea Association Resource Project, a multidisciplinary research consortium, began in 2007 to conduct a large-scale GWAS of Ansan and Ansung cohorts in the KoGES.26 From the 10 038 participants in both cohorts, we obtained genetic information of 10 004 individuals on microarrays, but we excluded data of 2011 individuals because they exhibited either a missing genotype call rate of <96% (n=401), heterozygosity <70% (n=11), gender consistencies (n=41), average pair-wise identity-by-state values higher than 0.8 as estimated values from first-degree relatives of Korean sib-pair samples (n=608) or medical history consisting of any kind of cancer (n=101). In addition, individuals who did not complete a pulmonary function test or anthropometric measurement (n=214), did not report smoking status (n=301) and reported a diagnosis of asthma (n=334) were omitted. The final GWAS consisted of 4319 individuals from Ansan and 3674 individuals from Ansung (see Supplementary Table S1).
Ethics statement
Informed written consent was obtained from all participants. The study protocol was approved by the ethics committee of the Korean Center for Disease Control and institutional review boards of the Korea University Ansan Hospital and the Ajou University School of Medicine.
Genotyping and quality control
For each subject, genomic DNA was separated from venous blood, and 500 ng of the sample was genotyped on the Affymetrix Genome-wide Human SNP array 5.0 (Affymetrix, Santa Clara, CA, USA). Bayesian robust linear modeling using the Mahalanobis distance genotyping algorithm was used for genotype calling of 500 568 SNPs.27 Detailed information on quality control method is available in a previous report.26 In Ansan, 312 381 SNP markers were used for the GWAS after excluding those with missing genotype call rates ⩾5% (n=45 343), deviations from the Hardy–Weinberg equilibrium test with statistical significance set at 0.0001 (n=35 410) or a minor allele frequency <5% (n=147 570). With the same criteria, final 313 984 SNP markers were included from Ansung.
GWA and haplotype analyses
To explore loci associated with pre-bronchodilator pulmonary function test values as a quantitative trait, we used the PLINK program, version 1.07 (Free Software Foundation, Boston, MA, USA) and performed multivariate linear regression models, including age, gender, site and height as covariates. Additive models were applied to the analysis and false discovery rate adjustments were used for multiple testing corrections.
We developed the linkage disequilibrium (LD) blocks using the Haploview program, version 4.2 (Broad Institute of Harvard and MIT, Cambridge, MA, USA). Lewontin’s |D′| and R2 between all pairs of biallelic SNP loci were used to examine LD using the default algorithm from Gabriel et al.28 We constructed the LD blocks for HapMap populations (available from the ver.3 HapMap project) using Haploview to cross-compare their LD patterns with those in our population. Haplotypes for the polymorphic SNPs were inferred with a minor allele frequency >0.10 and deviations from the Hardy–Weinberg equilibrium test with statistical significance set at 0.05. This procedure was also performed using the expectation-maximization algorithm that was implemented through the PLINK program.
Association of haplotypes with the value was analyzed using multivariate linear regression models including the same covariates, and similar analyses were conducted after stratification by smoking status (pack-years of smoking (PY)=0, 0
Results
Population characteristics
The average ages of the Ansan and Ansung cohorts were 48.99±7.82 and 55.60±8.74 years, respectively. Whereas 42.23% (n=1824) of the Ansan cohort participants were ‘ever-smokers’, 37.75% of the Ansung cohort participants were included in that category. Among all ‘ever smokers’, however, the average number of packs smoked per year was 21.05±16.05 in the Ansan cohort and 26.53±18.22 in the Ansung cohort. The pre-bronchodilator FEV1, FVC and FEV1/FVC values were significantly different between the two cohorts (Table 1).
Genome-wide association results with pulmonary function
To identify loci associated with FEV1/FVC, we conducted a multivariate linear regression analysis. The genomic inflation factor (λ) was estimated at 1.03 for FEV1/FVC in the combined set, suggesting no evidence of systematic bias. A Manhattan plot of GWAS from the combined set shows that SNPs associated with FEV1/FVC to the GWA significance level are located on chromosome 4q22.1 in the FAM13A gene (Figure 1). A quantile–quantile plot in Supplementary Figure S1 shows P-values for FEV1/FVC in −log10 scale across the diagonal line, but dots representing lower P-values at the right end of the plot are located above the line. For FEV1/FVC, significant GWA was seen for four SNPs in FAM13A, specifically on chromosome 4q22.1 after adjusting for P<0.05 using a false discovery rate control; P-values ranged from 1.76 × 10−7 to 5.47 × 10−7 in the combined set (8.38 × 10−6 to 2.61 × 10−5 in Ansan and 8.99 × 10−4 to 2.84 × 10−3 in Ansung). The most strongly associated SNP in this region, rs2609264 (P=1.76 × 10−7 in the combined set), is located in an intron of the FAM13A gene (Table 2 and Figure 2a).
Manhattan plot displaying results from genome-wide association of the pooled set. The pink dots shown in the FAM13A gene on 4q22.1 are associated with FEV1/FVC at a genome-wide significant level for the pooled set. The horizontal line indicates the genome-wide significance threshold P=1.00 × 10−5. FEV1, forced expiratory volume in the first second; FVC, forced vital capacity. A full color version of this figure is available at the Journal of Human Genetics journal online.
Regional association signals between single-nucleotide polymorphisms (SNPs) and FEV1/FVC and linkage disequilibrium (LD) within FAM13A. (a) Regional association plot between SNPs and FEV1/FVC within FAM13A. The association was drawn from multiple linear regression analysis adjusted for age, gender and pack-years of smoking on the basis of the additive model. (b) The LD among SNPs in FAM13A (from 89 868 514 to 90 193 745 bp) with the use of imputed genome-wide association (GWA) data. Within the block including the top four SNPs, the LD signal is very strong; the squared correlation coefficient between two loci (R2) was examined between 0.83 and 0.97. FEV1, forced expiratory volume in the first second; FVC, forced vital capacity. A full color version of this figure is available at the Journal of Human Genetics journal online.
For percent predicted FEV1 and percent predicted FVC, the GWA was seen for top 30 SNPs in multiple regions with P-values ⩽6.60 × 10−5 and 2.80 × 10−5, respectively (Supplementary Tables S3 and S4). The SNP with the smallest P-value for % predicted FEV1, rs825388 (P=3.85 × 10−6 in a combined set), is located on chromosome 5q14.3. Four SNPs on chromosome 5q14.3 near the GPR98 and two SNPs on chromosome 6p22.1 were significantly associated with % predicted FEV1. In addition, the most strongly association SNP for % predicted FVC, rs3131847 (P=1.65 × 10−6 in a combined set), is located on chromosome 6p22.1 near the ZFP57. Five SNPs on chromosome 6p22.1 near the ZFP57 and two SNPs on chromosome 4 in FAT1 were also significantly associated with % predicted FVC.
SNPs and haplotypes related to FEV1/FVC in the FAM13A locus
Focusing on FAM13A, we examined SNPs with LD in the association with FEV1/FVC in separate cohorts and their combined set. Using the additive model, four SNPs, rs2609264, rs1458551, rs2609261 and rs2609260, were found to have a strong LD. The squared correlation coefficient between two loci (R2) ranged from 0.83 to 0.97 in all parts. In addition, several intron SNPs in FAM13A, such as rs2869966, rs2869967 and rs2045517, were observed to be in high LD (R2=0.84) with our top SNP (rs2609264; Figure 2b). Previous large GWA studies demonstrated that three FAM13A SNPs are significantly, and in particular, associated with FEV1/FVC.11, 12
For the four SNPs in FAM13A, TCAG (43.60%) and CTGA (52.06%) were demonstrated to be the main haplotypes. In haplotype analysis, the CTGA haplotype had a highly significant association with lower FEV1/FVC than the TCAG haplotype (Table 3). The effect size for CTGA was −0.57% (s.e. =0.11, P=2.10 × 10−7) in the combined set. Compared with the TCAG haplotype, the CTGA haplotype was associated with lower FEV1/FVC by ∼0.75% (s.e.=0.17, P=9.83 × 10−6) in men and by ∼0.43% (s.e.=0.14, P=1.91 × 10−3) in women.
Associations of FAM13A haplotypes with FEV1/FVC by smoking behavior
Table 4 shows that TCAG and CTGA haplotypes are linked to significant differences in FEV1/FVC across all groups except for the mild smoking group (0
In addition, we found a statistically significant interaction between the CTGA haplotype and heavy smoking. In combined set, the CTGA haplotype in heavy smokers had significantly lower FEV1/FVC by 4.92% (s.e.=0.29, P=3.10 × 10−64) than would be expected if the two factors were independent: the effect size for CTGA haplotype compared with the TCAG haplotype on FEV1/FVC was −0.49% (s.e.=0.14, P=4.34 × 10−4) in nonsmokers; the effect size for heavy smoking status compared with nonsmoking on FEV1/FVC was −3.62% (s.e.=0.31, P=1.20 × 10−31) in TCAG haplotype group (P for interaction=0.028) (Figure 3 and Supplementary Table S2).
Joint effects of smoking behavior and haplotypes of FAM13A on reduced pulmonary function. Values on bars indicate reduced FEV1/FVC. Joint effects between smoking behavior and haplotype of FAM13A showed reduced FEV1/FVC; smokers with CTGA haplotype in heavy smokers had lower FEV1/FVC compared with TCAG haplotype in nonsmokers, and this effect is dose dependent on pack-years of smoking (PY) (after stratification by smoking status). In general linear model on contrast statement, P-value indicates significance for difference in haplotype distribution by smoking behavior. A full color version of this figure is available at the Journal of Human Genetics journal online.
Discussion
In this GWAS consisting of 7993 Korean participants, we identified GWA of FEV1/FVC with SNPs in FAM13A of 4q22.1: rs2609264, rs1458551, rs2609261 and rs2609260. For those four SNPs, the CTGA and the TCAG haplotypes were confirmed as main haplotypes. We found that the CTGA haplotype (‘risk haplotype’) was associated with lower FEV1/FVC than the TCAG haplotype (‘reference haplotype’). In addition, an interaction between the CTGA haplotype and heavy smoking was statistically significant.
Cigarettes include many oxidants, pro-oxidants and free radicals, all of which contribute to disturbed elasticity and extracellular matrix (ECM) of lung parenchyma.29 Existing literature also suggests that ECM-degrading enzymes, reactive oxygen species and airway inflammation correlate with smoke-related COPD.30, 31 In addition, Framingham Heart Study reported that cigarette smoking was strongly associated with a more rapid decline in pulmonary function, ∼0.2–0.3% decline in FEV1/FVC per year for each pack per day of smoking. This report also revealed that evidence for heritability is present in the rate of pulmonary impairment in family members; the heritability for decline in lung function ranged from 0.11 to 0.29 in nonsmokers, but from 0.14 to 0.39 in smokers.8 In summary, this study has shown evidence of a genetic contribution to pulmonary function decline, as well as a cigarette smoking behavior. In our study, the CTGA haplotype was associated with lower FEV1/FVC by ∼0.51% in nonsmokers and by ∼1.40% in heavy smokers, as compared with the TCAG haplotype. Our findings identified genetic associations about reduced pulmonary function in both nonsmokers and heavy smokers.
As stated above, current smoking status is the major environmental risk factor for the development of COPD, but some of the ex-smokers show continuing decrease in pulmonary function after they stop smoking.32 That is, the degree of structural change of airway is diverse among people, and genetic differences could be suggested as one of the causes. One study has reported that cigarette smoking not only affects the lung components, but its effects may also be modulated by genes of exposed individuals.33 We confirmed the joint effect of FAM13A gene and smoking status. The CTGA haplotype in heavy smokers had significantly lower FEV1/FVC than would be expected if the CTGA haplotype and heavy smoking were independent. In other words, our finding suggests that the risk of reduced pulmonary function is particularly high in FAM13A CTGA haplotype carriers who have heavy smoking status. Although several studies have examined the joint effect of smoking and some candidate genes for COPD, such as interleukin-6, interferon-γ, and α1-antitrypsin,34, 35, 36 none of the tests of interaction between smoking and FAM13A gene have been investigated in published GWAS of pulmonary function or COPD. We found significant gene-by-smoking interaction between FAM13A haplotypes and FEV1/FVC in heavy smokers.
Candidate genes that have been implicated in the pathogenesis of COPD may be involved in proteolysis/antiproteolysis (for example, SERPINA1, MMPs and SERPINA3), oxidants/antioxidants (for example, GST and SOD) and inflammation (for example, TNF-α and TGFβ).37, 38, 39, 40, 41, 42 Although pathophysiological mechanisms behind the association between the candidate genes and pulmonary function are still unclear, we identified FAM13A as one of the candidate genes for the related outcomes. A previous report suggested that FAM13A (or FAM13A1) may be associated with the ECM cluster that includes SPP1, MEPE, IBSP, DMP1 and DSPP. The ECM cluster genes reside within the same open chromatin domain, and expression of FAM13A and other ECM cluster genes may be coregulated.43 We observed that rs1585616 in the MEPE gene (an ECM gene) is weakly associated with FEV1/FVC, suggesting that potential biological mechanisms may bridge FAM13A and reduced lung function (data not shown). In addition, multiregional genes have been reported in large-scale GWASs (for example, DDX1, RARB, HTR4, ADAM19, GPR126, CDC123, LRP1 and THSD4).11, 15, 18, 44, 45 In our study, several SNPs of those candidate genes showed association with FEV1/FVC (Supplementary Table S5).
Herein, we describe the large-scale GWAS in an Asian population to identify susceptible genes related to decreased pulmonary function and compared their effects with environmental effects. In addition, this study is the first to report significant interaction between FAM13A gene and smoking behavior with a pulmonary function trait in Asians. Our study contains several limitations, however. First, some of the recent population-based GWASs reported that pulmonary function and/or COPD were associated with rs2045517, rs2869966, rs2869967, rs6830970 and rs7671167 in FAM13A, but we could not replicate the same results because these SNPs were not contained in our genetic data. Second, the biological function and pathophysiological mechanisms of FAM13A in humans is yet to be clarified. Therefore, fine-mapping within the FAM13A gene is required to confirm about association between statistically significant noncoding SNPs and expression of the FAM13A gene. Furthermore, biological pathway and network analysis using these GWAS results is needed to define a functional role for FAM13A in reduced pulmonary function or COPD.
In conclusion, our study reidentified that the FAM13A locus is related to pulmonary function in Asians. We also suggest that the FAM13A variants may modify the adverse effects of cigarette smoking in lower pulmonary function, especially in heavy smokers. To clarify furthermore the interaction of FAM13A gene or other genes with environmental exposures on reduced lung function, functional studies may be necessary in the future.
References
Barreiro, T. J . & Perillo, I . An approach to interpreting spirometry. Am. Fam. Physician. 69, 1107–1114 (2004).
Rabe, K. F ., Hurd, S ., Anzueto, A ., Barnes, P. J ., Buist, S. A ., Calverley, P . et al. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease: GOLD executive summary. Am. J. Respir. Crit. Care Med. 176, 532–555 (2007).
Lopez, A. D ., Mathers, C. D ., Ezzati, M ., Murray, C. J. L . & Jamison, D. T . Global Burden of Disease and Risk Factors, (The World Bank and Oxford University Press, New York, USA, 2006).
Schünemann, H. J ., Dorn, J ., Grant, B. J ., Winkelstein, W. Jr & Trevisan, M . Pulmonary function is a long-term predictor of mortality in the general population: 29-year follow-up of the Buffalo Health Study. Chest 118, 656–664 (2000).
Pellegrino, R ., Viegi, G ., Brusasco, V ., Crapo, R. O ., Burgos, F ., Casaburi, R . et al. Interpretative strategies for lung function tests. Eur. Respir. J. 26, 948–968 (2005).
Mannino, D. M . & Buist, A. S . Global burden of COPD: risk factors, prevalence, and future trends. Lancet 370, 765–773 (2007).
Snider, G. L . Chronic obstructive pulmonary disease: risk factors, pathophysiology and pathogenesis. Annu. Rev. Med 40, 411–429 (1989).
Gottlieb, D. J ., Wilk, J. B ., Harmon, M ., Evans, J. C ., Joost, O ., Levy, D . et al. Heritability of longitudinal change in lung function. The Framingham study. Am. J. Respir. Crit. Care Med. 164, 1655–1659 (2001).
Hemminki, K ., Li, X ., Sundquist, K . & Sundquist, J . Familial risks for chronic obstructive pulmonary disease among siblings based on hospitalisations in Sweden. J. Epidemiol. Community Health. 62, 398–401 (2008).
Seifart, C . & Plagens, A . Genetics of chronic obstructive pulmonary disease. Int. J. Chron. Obstruct. Pulmon. Dis. 2, 541–550 (2007).
Hancock, D. B ., Eijgelsheim, M ., Wilk, J. B ., Gharib, S. A ., Loehr, L. R ., Marciante, K. D . et al. Meta-analyses of genome-wide association studies identify multiple loci associated with pulmonary function. Nat. Genet. 42, 45–52 (2010).
Cho, M. H ., Boutaoui, N ., Klanderman, B. J ., Sylvia, J. S ., Ziniti, J. P ., Hersh, C. P . et al. Variants in FAM13A are associated with chronic obstructive pulmonary disease. Nat. Genet. 42, 200–202 (2010).
Pillai, S. G ., Kong, X ., Edwards, L. D ., Cho, M. H ., Anderson, W. H ., Coxson, H. O . et al. Loci identified by genome-wide association studies influence different disease-related phenotypes in COPD. Am. J. Respir. Crit. Care Med. 182, 1498–1505 (2010).
Wilk, J. B ., Chen, T. H ., Gottlieb, D. J ., Walter, R. E ., Nagle, M. W ., Brandler, B. J . et al. A genome-wide association study of pulmonary function measures in the Framingham Heart Study. PLoS Genet. 5, e1000429 (2009).
Repapi, E ., Sayers, I ., Wain, L. V ., Burton, P. R ., Johnson, T ., Obeidat, M . et al. Genome-wide association study identifies five loci associated with lung function. Nat. Genet. 42, 36–44 (2010).
Cho, M. H ., Castaldi, P. J ., Wan, E. S ., Siedlinski, M ., Hersh, C. P ., Demeo, D. L . et al. A genome-wide association study of COPD identifies as susceptibility locus on chromosome 19q13. Hum. Mol. Genet. 21, 947–957 (2012).
Soler Artigas, M ., Wain, L. V ., Repapi, E ., Obeidat, M ., Sayers, I ., Burton, P. R . et al. Effect of five genetic variants associated with lung function on the risk of chronic obstructive lung disease, and their joint effects on lung function. Am. J. Respir. Crit. Care Med. 184, 786–795 (2011).
Soler Artigas, M ., Loth, D. W ., Wain, L. V ., Gharib, S. A ., Obeidat, M ., Tang, W . et al. Genome-wide association and large-scale follow up identifies 16 new loci influencing lung function. Nat. Genet. 43, 1082–1090 (2011).
Kim, W. J ., Lee, M. K ., Shin, C ., Cho, N. H ., Lee, S. D ., Oh, Y. M . et al. Genome-wide association studies identify locus on 6p21 influencing lung function in the Korean population. Respirology 19, 360–368 (2014).
Kim, W. J ., Lim, M. N ., Hong, Y ., Silverman, E. K ., Lee, J. H ., Jung, B. H . et al. Association of lung function genes with chronic obstructive pulmonary disease. Lung 192, 473–480 (2014).
Guo, Y ., Lin, H ., Gao, K ., Xu, H ., Deng, X ., Zhang, Q . et al. Genetic analysis of IREB2, FAM13A and XRCC5 variants in Chinese Han patients with chronic obstructive pulmonary disease. Biochem. Biophys. Res. Commun. 415, 284–287 (2011).
Shin, C ., Kim, J ., Kim, J ., Lee, S ., Shim, J . & In, K . et al. Association of habitual snoring with glucose and insulin metabolism in nonobese Korean adult men. Am. J. Respir. Crit. Care Med. 171, 287–291 (2005).
Baik, I ., Kim, J ., Abbott, R. D ., Joo, S ., Jung, K ., Lee, S . et al. Association of snoring with chronic bronchitis. Arch. Intern. Med. 168, 167–173 (2008).
Cho, N. H ., Chan, J. C ., Jang, H. C ., Lim, S ., Kim, H. L . & Choi, S. H . Cigarette smoking is an independent risk factor for type 2 diabetes: a four-year community-based prospective study. Clin. Endocrinol. 71, 679–685 (2009).
American Thoracic Society Standardization of Spirometry, 1994 Update. Am. J. Respir. Crit. Care Med. 152, 1107–1136 (1995).
Cho, Y. S ., Go, M. J ., Kim, Y. J ., Heo, J. Y ., Oh, J. H ., Ban, H. J . et al. A large-scale genome-wide association study of Asian populations uncovers genetic factors influencing eight quantitative traits. Nat. Genet. 41, 527–534 (2009).
Rabbee, N . & Speed, T. P . A genotype calling algorithm for Affymetrix SNP arrays. Bioinformatics 22, 7–12 (2006).
Gabriel, S. B ., Schaffner, S. F ., Nguyen, H ., Moore, J. M ., Roy, J ., Blumenstiel, B . et al. The structure of haplotype blocks in the human genome. Science 296, 2225–2229 (2002).
Chow, C. K . Cigarette smoking and oxidative damage in the lung. Ann. N.Y. Acad. Sci. 686, 289–298 (1993).
Barnes, P. J ., Shapiro, S. D . & Pauwels, R. A . Chronic obstructive pulmonary disease: molecular and cellular mechanisms. Eur. Respir. J. 22, 672–688 (2003).
Zandvoort, A ., Postma, D. S ., Jonker, M. R ., Noordhoek, J. A ., Vos, J. T . & Timens, W . Smad gene expression in pulmonary fibroblasts: indications for defective ECM repair in COPD. Respir. Res. 9, 83 (2008).
Makris, D ., Moschandreas, J ., Damianaki, A ., Ntaoukakis, E ., Siafakas, N. M ., Milic Emili, J . et al. Exacerbations and lung function decline in COPD: new insights in current and ex-smokers. Respir. Med. 101, 1305–1312 (2007).
Conrad, B . & Antonarakis, S. E . Gene duplication: a drive for phenotypic diversity and cause of human disease. Annu. Rev. Genomics Hum. Genet. 8, 17–35 (2007).
Hackett, T. L ., Stefanowicz, D ., Aminuddin, F ., Sin, D. D ., Connett, J. E ., Anthonisen, N. R . et al. Effect of gene environment interactions on lung function and cardiovascular disease in COPD. Int. J. Chron. Obstruct. Pulmon. Dis. 6, 277–287 (2011).
Sadeghnejad, A ., Meyers, D. A ., Bottai, M ., Sterling, D. A ., Bleecker, E. R . & Ohar, J. A . IL13 promoter polymorphism 1112C/T modulates the adverse effect of tobacco smoking on lung function. Am. J. Respir. Crit. Care Med. 176, 748–752 (2007).
Castaldi, P. J ., Demeo, D. L ., Hersh, C. P ., Lomas, D. A ., Soerheim, I. C ., Gulsvik, A . et al. Impact of non-linear smoking effects on the identification of gene-by-smoking interactions in COPD genetics studies. Thorax 66, 903–909 (2011).
Stoller, J. K . & Aboussouan, L. S . Alpha1-antitrypsin deficiency. Lancet 365, 2225–2236 (2005).
Joos, L ., He, J. Q ., Shepherdson, M. B ., Connett, J. E ., Anthonisen, N. R ., Paré, P. D . et al. The role of matrix metalloproteinase polymorphisms in the rate of decline in lung function. Hum. Mol. Genet 11, 569–576 (2002).
He, J. Q ., Ruan, J ., Connett, J. E ., Anthonisen, N. R ., Paré, P. D . & Sandford, A. J . Antioxidant gene polymorphisms and susceptibility to a rapid decline in lung function in smokers. Am. J. Respir. Crit. Care Med. 166, 323–328 (2002).
Juul, K ., Tybjaerg-Hansen, A ., Marklund, S ., Lange, P . & Nordestgaard, B. G . Genetically increased antioxidative protection and decreased chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 173, 858–864 (2006).
Sakao, S ., Tatsumi, K ., Igari, H ., Shino, Y ., Shirasawa, H . & Kuriyama, T . Association of tumor necrosis factor alpha gene promoter polymorphism with the presence of chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 163, 420–422 (2001).
Celedón, J. C ., Lange, C ., Raby, B. A ., Litonjua, A. A ., Palmer, L. J ., DeMeo, D. L . et al. The transforming growth factor-beta1 (TGFB1) gene is associated with chronic obstructive pulmonary disease (COPD). Hum. Mol. Genet. 13, 1649–1656 (2004).
Cohen, M ., Reichenstein, M ., Everts-van der Wind, A ., Heon-Lee, J ., Shani, M ., Lewin, H. A . et al. Cloning and characterization of FAM13A1—a gene near a milk protein QTL on BTA6: evidence for population-wide linkage disequilibrium in Israeli Holsteins. Genomics 84, 374–383 (2004).
Hancock, D. B ., Artigas, M. S ., Gharib, S. A ., Henry, A ., Manichaikul, A ., Ramasamy, A . et al. Genome-wide joint meta-analysis of SNP and SNP-by-smoking interaction identifies novel loci for pulmonary function. PLoS Genet. 8, e1003098 (2012).
Yao, T. C ., Du, G ., Han, L ., Sun, Y ., Hu, D ., Yang, J. J . et al. Genome-wide association study of lung function phenotypes in a founder population. J Allergy Clin Immunol 133, 248–255 e1-10 (2014).
Acknowledgements
This study was supported by the Korea Centers for Disease Control and Prevention, Ministry for Health, Welfare and Family Affairs, Republic of Korea (Grants 2001-347-6111-221 and 2002-347-6111-221, and the Korean Genome Analysis Project (4845-301)).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Kim, S., Kim, H., Cho, N. et al. Identification of FAM13A gene associated with the ratio of FEV1 to FVC in Korean population by genome-wide association studies including gene–environment interactions. J Hum Genet 60, 139–145 (2015). https://doi.org/10.1038/jhg.2014.118
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2014.118